When you convert a JPEG to a DICOM file, you’re doing much more than just changing the file extension. You're fundamentally transforming a simple picture into a clinically rich piece of medical data, embedding it within the robust structure of the DICOM standard. It's a critical upgrade that ensures the image can talk to medical systems like PACS and EHRs, maintaining data integrity and diagnostic accuracy along the way.
Why Convert JPEGs to DICOM for Medical Imaging?
In modern healthcare, turning a basic JPEG into a DICOM object is an essential task. A standard JPEG file is really just a collection of pixels—it's a picture, and that's it. A DICOM file, on the other hand, is an image and a detailed patient record all rolled into one. It contains a rich header packed with metadata that gives the image vital clinical context.
This distinction is what really matters. Let’s say a dermatologist snaps a photo of a skin lesion with a digital camera. As a JPEG, it's just a standalone picture. For that image to have any real value inside a hospital's network, it needs to be linked to a specific patient, a particular study, and a clinical encounter. That’s exactly what the DICOM structure is designed to do.
Bridging the Gap Between Pictures and Patient Data
At its core, converting a JPEG to DICOM is all about integrating an isolated image into the larger clinical workflow. Healthcare systems are built around the concept of interoperability, and the DICOM standard is the universal language that imaging devices, PACS (Picture Archiving and Communication System), and EHRs (Electronic Health Records) all speak.
Without this conversion, a JPEG from an outside source is just an outlier. It's incredibly difficult to archive, track, or properly associate with a patient's medical history. The conversion process enriches the raw image with all the necessary data tags, including:
- Patient Demographics (Name, ID, Date of Birth)
- Study Information (Study Date, Referring Physician)
- Equipment Details (Modality, Manufacturer)
- Series and Image Identifiers (Unique UIDs for traceability)
The upgrade from JPEG to DICOM isn't just a technicality. It’s about elevating a simple image into a compliant, data-rich asset that supports diagnostic integrity and seamless clinical operations.
Key Differences Between JPEG and DICOM Files
To really appreciate the "why" behind the conversion, it helps to see a direct comparison. It quickly becomes clear why a basic JPEG just doesn't cut it for clinical purposes. The DICOM format was built from the ground up to handle the complex demands of medical diagnostics and record-keeping.
| Feature | JPEG | DICOM |
|---|---|---|
| Metadata | Minimal (camera settings, date). | Extensive (patient info, study details, equipment data, unique IDs). |
| Context | Standalone image with no clinical context. | Integrated object within a patient’s full medical record. |
| Interoperability | Not natively compatible with PACS or EHRs. | The universal standard for medical imaging systems. |
| Traceability | Lacks unique identifiers for tracking. | Uses globally unique identifiers (UIDs) for every study, series, and image. |
| Use Case | General-purpose photography. | Regulated medical diagnostics and clinical workflows. |
This table shows that the conversion is driven by the need for a file structure that can carry much more than just pixel data.
The process of converting JPEGs into the DICOM format is a cornerstone of modern medical imaging. As you can see, it’s not just about preserving image quality; it’s about enriching the file with the necessary patient and study metadata to support better clinical workflows. You can learn more about how this integration boosts diagnostic confidence from PostDICOM's comprehensive guide.
Getting this conversion right is a non-negotiable skill in healthcare IT. It sets the stage for the practical, hands-on coding examples we'll dive into next.
Alright, before we get into the nitty-gritty of converting JPEGs to DICOM files, we need to get our workspace in order. A clean, well-organized Python environment is non-negotiable. Trust me, spending a few minutes on setup now saves you from hours of frustrating dependency headaches later.
First things first, you'll need a modern version of Python. If you don't already have it, you can grab the latest stable release from the official Python website. During the installation, there's a little checkbox that says "Add Python to PATH"—make sure you tick that one. It's a small detail that makes running Python from your command line a whole lot easier.
Getting the Right Tools for the Job
With Python installed, we need to bring in a couple of specialized libraries. These are the workhorses that will handle the heavy lifting.
- Pydicom: This is the gold standard for working with DICOM files in Python. It lets you read, modify, and write DICOM data with incredible flexibility.
- Pillow: A modern fork of the classic Python Imaging Library (PIL), Pillow is what we'll use to actually read the pixel data from our JPEG images.
Installing them is a breeze using pip, Python's built-in package manager. Just open up your terminal or command prompt and run these two commands one after the other:
pip install pydicom
pip install Pillow
And that’s it. These commands fetch the latest versions of both libraries and make them ready to use in your scripts.
A Quick Tip from the Trenches: If you're doing any serious development, always work inside a virtual environment. It's a simple practice that isolates your project's libraries from the rest of your system, preventing version conflicts that can be a nightmare to debug.
Creating one is straightforward. Just navigate to your project directory in the terminal and type:
python -m venv my_dicom_env
Then, you just need to activate it before you install anything. On Windows, you'll run my_dicom_env\Scripts\activate. On a Mac or Linux machine, it's source my_dicom_env/bin/activate. Making this a habit will keep your jpeg to dicom conversion project tidy and easily shareable.
Building Your First JPEG to DICOM Converter
Okay, with the environment set up, let's get our hands dirty and build a foundational Python script. We're going from theory to practice, creating a simple but functional converter that takes a JPEG and turns it into a DICOM object. The idea is to build something you can understand and then expand on later.
Our game plan is straightforward: we'll use Pillow to read the JPEG file, then lean on Pydicom to create a brand new DICOM dataset from scratch. The most important part is populating that dataset with the critical metadata that makes it a valid DICOM file. This isn't just a copy-paste exercise; it's about understanding how to construct a DICOM object that a PACS will actually recognize and accept.
The Core Logic of the Converter Script
At its heart, the script does three main things. First, it pulls in the pixel data from your source JPEG. Then, it builds a DICOM dataset—think of it as a special kind of dictionary where the keys are DICOM tags and the values are the data. Finally, it saves this newly created dataset as a .dcm file.
This diagram lays out the high-level workflow into three distinct stages.
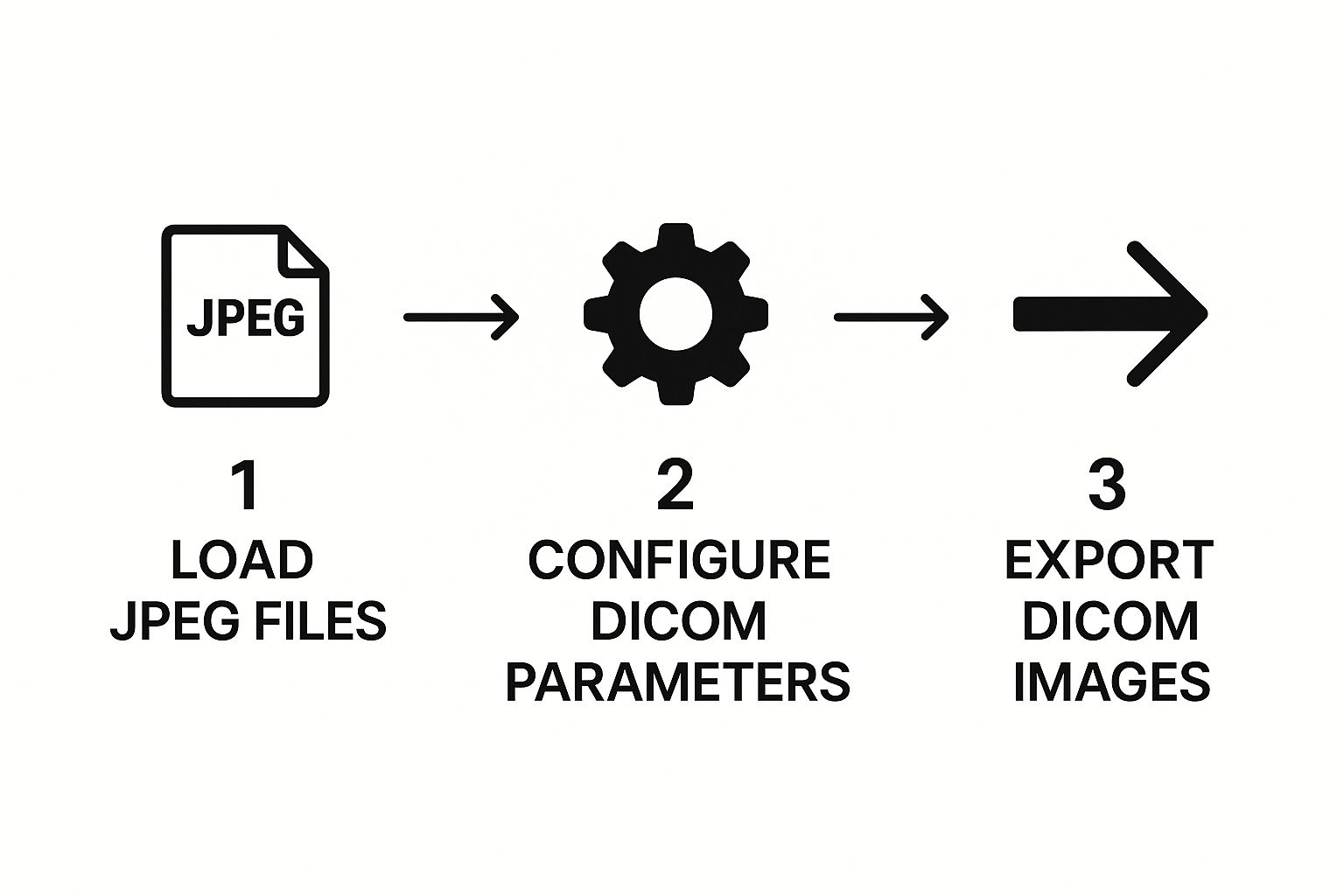
As you can see, the process flows from simply loading the image to configuring the essential medical metadata, and finally, exporting the complete DICOM file.
Crafting the Python Code
First things first, let's import the libraries we need and define our file paths. We'll need pydicom for all the DICOM heavy lifting, Pillow (as PIL) for handling the image, and Python's built-in datetime and os modules for things like timestamps and file management.
import pydicom
from pydicom.dataset import Dataset, FileDataset
from pydicom.uid import generate_uid
from PIL import Image
import datetime
import os
Define input JPEG and output DICOM file paths
jpeg_file_path = 'path/to/your/image.jpg'
dicom_file_path = 'path/to/your/output.dcm'
Load the JPEG image using Pillow
try:
jpeg_image = Image.open(jpeg_file_path)
except FileNotFoundError:
print(f"Error: The file {jpeg_file_path} was not found.")
exit()
This initial chunk of code takes care of loading the file and includes a basic error check if the file doesn't exist—a good habit for any robust script. As you build this out, remember that following principles of how to write clean code will make your life much easier when you need to maintain or scale your solution.
Next up, we'll create the DICOM dataset and start filling it with the most critical Type 1 tags. These are the non-negotiable pieces of metadata required for a file to be considered valid.
The heart of a DICOM file is its metadata. Without correctly generated Unique Identifiers (UIDs) for the study, series, and instance, a PACS will almost certainly reject the file. Pydicom's
generate_uid()function is the perfect tool for this.
Here’s how you can add this crucial information:
Create a new DICOM dataset
ds = Dataset()
Set patient and study information
ds.PatientName = "Test^Patient"
ds.PatientID = "123456"
ds.StudyDate = datetime.date.today().strftime('%Y%m%d')
ds.StudyTime = datetime.datetime.now().strftime('%H%M%S')
Generate and add UIDs
ds.StudyInstanceUID = generate_uid()
ds.SeriesInstanceUID = generate_uid()
ds.SOPInstanceUID = generate_uid()
Every time you call generate_uid(), you get a new, globally unique identifier. This is what ensures every image, series, and study can be tracked without ambiguity across any healthcare system.
Embedding the Pixel Data
The final piece of the puzzle is getting the actual image data from our JPEG into the DICOM structure. This involves more than just a simple copy; we have to set specific tags that describe the image itself, like its dimensions, color space, and compression method.
A few key tags to set are:
- Photometric Interpretation: This tells a DICOM viewer how to interpret the color data. For a standard color JPEG, the value is typically
YBR_FULL_422. - Samples per Pixel: For an RGB-style image, this value is 3.
- Pixel Representation: We use 0 here, which indicates the pixel data is unsigned.
Here is the code that handles embedding the pixel data and saving the final file:
Set image-specific metadata
ds.Rows, ds.Columns = jpeg_image.height, jpeg_image.width
ds.PhotometricInterpretation = "YBR_FULL_422"
ds.SamplesPerPixel = 3
ds.BitsAllocated = 8
ds.BitsStored = 8
ds.HighBit = 7
ds.PixelRepresentation = 0
ds.Modality = "OT" # Other
Embed the pixel data
ds.PixelData = jpeg_image.tobytes()
Create a FileDataset to save
file_meta = Dataset()
file_meta.MediaStorageSOPClassUID = '1.2.840.10008.5.1.4.1.1.7' # Secondary Capture Image Storage
file_meta.MediaStorageSOPInstanceUID = ds.SOPInstanceUID
file_meta.ImplementationClassUID = pydicom.uid.pydicom_implementation_class_uid
file_meta.TransferSyntaxUID = pydicom.uid.ImplicitVRLittleEndian
output_dataset = FileDataset(dicom_file_path, {}, file_meta=file_meta, preamble=b"\0" * 128)
output_dataset.update(ds)
output_dataset.save_as(dicom_file_path, write_like_original=False)
print(f"Successfully converted {jpeg_file_path} to {dicom_file_path}")
And there you have it. This script is a complete, working example of a jpeg to dicom conversion. It provides a solid foundation you can build on for more advanced features.
Getting DICOM Metadata and Compliance Right
A DICOM file is so much more than just a picture with a .dcm extension. Without the right metadata, that's all it is. The real clinical power comes from the standardized data header, which transforms a simple image into a traceable, diagnostic-quality record that a PACS can actually understand, archive, and display correctly.
Getting this part right isn’t optional; it's a hard requirement. If you botch the metadata—by leaving out critical tags or putting in junk data—a PACS will almost certainly reject your file. That can bring a clinical workflow to a grinding halt.
Navigating Mandatory and Optional DICOM Tags
The DICOM standard is very specific about what data it needs. Tags are categorized into different types to make sure every file has a consistent baseline of information. Here’s how it breaks down:
- Type 1 (Mandatory): These are the non-negotiables. You must include these tags, and they must have a valid value. Think
PatientID,StudyInstanceUID, andSOPClassUID. Leave one of these out, and your file is dead on arrival. - Type 2 (Required): You have to include the tag itself, but its value can be empty if the information simply isn't known or doesn't apply. A good example is
ReferringPhysicianName. The tag must be there, but the value can be null if you don't have it. - Type 3 (Optional): Just like it sounds. You can add these for extra context if you have the data, but it’s no big deal if you leave them out.
PatientCommentsis a classic Type 3 tag.
My advice? When you're starting a conversion project, tackle all the Type 1 requirements first. They're the absolute foundation of a valid DICOM file. Once you've got those locked in, you can move on to the Type 2 and any useful Type 3 tags to add more detail.
When you convert a JPEG, you’re usually creating what's called a "secondary capture." This means the image didn't come directly from a modality like a CT or MRI scanner. To reflect this, you’ll need to set the Modality tag specifically to "OT" (for "Other").
Essential DICOM Tags for a Valid Conversion
To make a JPEG truly useful in a clinical setting, you need to embed a core set of metadata. The table below outlines some of the most critical tags you'll need to populate for a typical secondary capture. This isn't an exhaustive list, but it's the bare minimum you'll need to get right.
| DICOM Tag Name | Tag ID | Requirement Type | Description & Example |
|---|---|---|---|
| Patient ID | (0010,0020) | Type 1 | The unique identifier for the patient within the institution. Example: 12345678 |
| Patient Name | (0010,0010) | Type 2 | The full name of the patient. Example: Doe^John |
| Study Instance UID | (0020,000D) | Type 1 | A globally unique ID for the entire study. Example: 1.2.826.0.1... (generated) |
| Series Instance UID | (0020,000E) | Type 1 | A globally unique ID for this specific series of images. Example: 1.2.826.0.1... (generated) |
| SOP Instance UID | (0008,0018) | Type 1 | A globally unique ID for this specific image instance. Example: 1.2.826.0.1... (generated) |
| SOP Class UID | (0008,0016) | Type 1 | Defines the type of DICOM object. Example: 1.2.840.10008.5.1.4.1.1.7 (for Secondary Capture) |
| Modality | (0008,0060) | Type 1 | The type of acquisition. For JPEGs, this is "OT". Example: OT |
| Study Date | (0008,0020) | Type 2 | The date the study was performed. Format: YYYYMMDD |
| Series Number | (0020,0011) | Type 2 | A number that orders series within a study. Example: 1 |
| Instance Number | (0020,0013) | Type 2 | A number that orders instances within a series. Example: 1 |
Getting these tags populated correctly is the difference between a file that works and one that gets rejected.
The Critical Role of UIDs and Series Consistency
I can't stress this enough: the Unique Identifiers (UIDs) are probably the most important pieces of metadata you'll generate. Every single study, series, and image instance in the DICOM universe needs its own globally unique ID.
Imagine you're converting a set of photos of a skin lesion taken a few minutes apart. These images all belong together. To link them, they must all share the same StudyInstanceUID and SeriesInstanceUID.
At the same time, each individual image is its own unique object. That means every single file needs its own distinct SOPInstanceUID. This hierarchy is precisely what lets a PACS group the images properly into a single, cohesive series. Using a library like Pydicom to generate these UIDs is the standard way to handle this.
Ensuring Compliance and Data Privacy
Beyond just getting the file structure right, you have to think about compliance. The rise of jpeg to dicom conversion tools is driven by the need for better interoperability, but it comes with serious responsibility. You're handling Protected Health Information (PHI).
That means your conversion script needs to be built with patient privacy in mind, adhering to regulations like HIPAA in the U.S. or GDPR in Europe. If you're interested in how modern platforms handle this balance, you can explore detailed insights on DICOM conversion tools that are built for clinical environments.
Ultimately, your conversion process must include steps for securely handling patient data. If the images are ever going to be used for research or teaching, you'll need a reliable anonymization workflow. A compliant, secure process is what makes your converted JPEG a truly valid and useful clinical asset.
Fine-Tuning Your Conversion Workflow
Once you have a basic script that can convert a single JPEG, the real work begins. Moving from a simple, one-off converter to a professional, resilient tool means thinking about efficiency, handling inevitable errors, and optimizing for real-world use. This is what separates a functional script from a production-ready solution for your jpeg to dicom conversion needs.
One of the first things you'll want to tackle is file size. While your original JPEG is already compressed, DICOM has its own set of internal compression schemes, known as Transfer Syntaxes. Choosing the right one is always a balancing act between minimizing storage space and preserving image quality.
Choosing the Right Compression and Transfer Syntax
A fantastic option to explore here is JPEG 2000. Its integration into the DICOM standard was a big step forward, offering a powerful combination of efficient compression and high diagnostic image quality. Unlike the standard JPEG you're starting with, JPEG 2000 supports both lossless and lossy modes, giving you much finer control. If you want to dig deeper into its role in medical imaging, Minnovaa has a detailed overview worth reading.
Putting this into practice with Pydicom is surprisingly simple. All you need to do is specify the TransferSyntaxUID when you save the file. For example, to use JPEG 2000 Lossless, you'd set the UID to '1.2.840.10008.1.2.4.90'. This one small change can dramatically shrink your file sizes without compromising the critical details needed for diagnosis.
Building a Bulletproof Batch Processor
Converting a single file is a good start, but let's be realistic—you’re probably dealing with entire folders of images. A batch processing function is a must-have to automate the work and save a ton of time. The core logic is straightforward: just loop through a directory, find all the .jpg files, and feed each one into your conversion function.
But a truly robust script has to anticipate that things will go wrong. What if a file is corrupted? Or what if a non-image file somehow ended up with a .jpg extension? This is where solid error handling makes all the difference.
Nothing is more frustrating than a script that crashes halfway through a large batch. By wrapping your main conversion logic in a
try...exceptblock, you can gracefully handle errors on a per-file basis, log the problem, and keep the process running for the rest of the folder.
This simple addition prevents one bad apple from spoiling the whole bunch. For instance, you can catch a FileNotFoundError or an UnidentifiedImageError from the Pillow library, print a helpful message to your log, and just move on to the next file.
Using Logging and Metadata Templates
For any serious workflow, especially in a clinical or research environment, you need an audit trail. Instead of just printing messages to the console where they can get lost, use Python's built-in logging module. This lets you write status updates, errors, and success confirmations to a dedicated log file, which is invaluable for traceability and debugging.
A good log should capture key details for each file:
- The timestamp of the conversion attempt.
- The original source JPEG filename.
- The new DICOM filename and its unique identifiers (UIDs).
- A clear message for any errors that occurred.
To make batch processing even smoother, think about creating a metadata template. You can define a base Dataset object with all the static information already filled out—things like StudyDate or Modality that won't change between images in the same batch. For each new JPEG, you just copy this template and add the instance-specific details like SOPInstanceUID and the pixel data.
This approach drastically cuts down on repetitive code and, more importantly, reduces the risk of typos or other manual data entry mistakes. It makes your entire jpeg to dicom conversion process faster, more consistent, and far more reliable.
Common Questions About JPEG to DICOM Conversion

As you get your hands dirty with medical imaging, you'll find a few questions about JPEG to DICOM conversion come up again and again. Getting these sorted out is the first step toward building a solid, reliable workflow that produces images ready for clinical use.
A common one I hear is whether any old JPEG can be turned into a DICOM file. While technically you can wrap almost any image, the real question is whether you should. A DICOM file without accurate, rich medical metadata is essentially useless in a clinical setting. A PACS will almost certainly reject a file that's missing the necessary patient and study details.
Another big worry is about image quality. People often ask if the conversion itself will mess up the original picture.
Will I Lose Image Quality During the Conversion?
This is a really common misconception, but the good news is the answer is no. The conversion process itself doesn't actually touch the pixel data. It's more like taking your original JPEG pixel stream and carefully placing it inside the larger DICOM structure. The visual quality you started with is exactly what you end up with.
The only way you'd see quality loss is if you chose to re-compress the image with another lossy algorithm while creating the DICOM file. To maintain diagnostic integrity, the best practice is always to embed the original JPEG data as-is, without adding another layer of compression.
Your original pixel data is preserved. Think of it like putting a photo into a detailed, labeled envelope—the photo inside doesn't change, but it gains a wealth of contextual information that makes it clinically valuable.
Finally, you absolutely have to get your head around DICOM UIDs. These Unique Identifiers are the foundation of traceability in the medical imaging world. Without them, everything falls apart.
- Study Instance UID: This links all the different images from a single medical exam together.
- Series Instance UID: This groups related images within that study, like all the axial slices.
- SOP Instance UID: This is the unique ID for each individual image file.
If you generate these UIDs incorrectly or, even worse, duplicate them, you're setting yourself up for major problems. It can lead to disastrous data mix-ups or cause the receiving system to reject your files outright. Always use a tried-and-true method, like Pydicom's generate_uid() function, to make sure every identifier is truly unique.
At PYCAD, we specialize in building the intelligent systems that power modern medical imaging. If you're looking to integrate advanced AI capabilities into your diagnostic workflows, explore our services at https://pycad.co.






