At its core, a sample DICOM file is a non-production medical imaging file created specifically for things like software testing, research, and education. It's built just like a real one, containing both the pixel data (the image) and a metadata header, but with one crucial difference: all the patient and equipment info is anonymized. This makes it completely safe to use without ever touching real patient data.
Understanding Why You Need A Sample Dicom File
Before you even start looking for a sample DICOM file, it helps to get a feel for what it actually is and why it's so fundamental to medical imaging. A DICOM file isn't just a picture; it's a complete, self-contained data package. That's what makes it the gold standard.
Think of it as having two distinct, but connected, parts:
- Pixel Data: This is the visual part—the actual grayscale or color image that comes off a CT scanner, MRI, or X-ray machine. It's the raw diagnostic information a radiologist would analyze.
- Metadata Header: This is a text-based section loaded with information, organized into "tags," that describe everything you could possibly want to know about the image. It includes patient demographics (anonymized, of course), details about the study, and even the technical specs of the machine that took the scan.
This all-in-one format solved a huge problem back in the day. The DICOM (Digital Imaging and Communications in Medicine) standard was born in the early 1980s out of pure necessity. Images from one manufacturer's scanner simply wouldn't work with another's software. A joint committee from the American College of Radiology (ACR) and the National Electrical Manufacturers Association (NEMA) stepped in to create the standard we rely on today, ensuring that any compliant software can read and correctly interpret any DICOM file. If you're curious, you can get the full history of DICOM on its Wikipedia page.
Real-World Applications
So, why are these sample files so important? Well, using actual patient data for development or research is a minefield of ethical and legal issues, all governed by strict regulations like HIPAA. Sample files give you a safe and effective way to work around that.
Imagine you're a software developer building a new PACS (Picture Archiving and Communication System) viewer. You need to test your application with everything you can throw at it. That means you need a wide variety of datasets—from simple, single-slice X-rays to complex, multi-frame cardiac ultrasounds—to make sure your viewer can handle different modalities and render images perfectly. Using samples lets you do all that without ever touching protected health information.
Or, maybe you're a data scientist training an AI model to spot anomalies in brain scans. You're going to need thousands of images to build a reliable algorithm. A large repository of anonymized sample DICOM files is the only ethical way to get the volume of data you need to build and validate your model.
A well-curated set of sample DICOM files is invaluable for many different professionals. The table below breaks down some of the most common scenarios where these files are put to use.
Common Use Cases for Sample DICOM Files
| Use Case | Primary User | Key Requirement |
|---|---|---|
| Software Development & Testing | PACS/Viewer Developers | Diverse modalities, various vendors, edge cases (e.g., corrupted headers) |
| AI & Machine Learning | Data Scientists, Researchers | Large, labeled datasets, specific pathologies or anatomical regions |
| Training & Education | Medical Students, Radiologists in Training | Classic examples of specific conditions, high-quality images for learning |
| System Interoperability | Healthcare IT Professionals | Files from different manufacturers to test connectivity and data exchange |
| Sales & Product Demos | Medical Device Sales Teams | Visually impressive, clean data that showcases software/hardware features |
Whether you're debugging code, training a neural network, or demonstrating a new product, having the right kind of sample data is often the first step to success.
Key Takeaway: Sample DICOM files are essential tools for driving innovation and education in medical technology. They give developers, researchers, and students a way to work with realistic imaging data in a secure, compliant, and controlled environment, which helps speed up progress without putting patient privacy at risk.
Hunting Down Public DICOM Data
So, you need to get your hands on some sample DICOM files. Whether you're building a new viewer, testing an anonymization script, or training a machine learning model, you'll need good data. Fortunately, you don't have to start from scratch. There are some fantastic public repositories out there, packed with anonymized data just waiting to be used.
The trick is knowing where to look. Each archive has its own personality and purpose. Some are massive, research-focused behemoths, while others are geared more toward providing clean, simple examples for developers. Let's dig into a few of my go-to sources.
The Cancer Imaging Archive (TCIA)
If your work touches anything related to oncology, your first stop should absolutely be The Cancer Imaging Archive (TCIA). This is the big one. Funded by the National Cancer Institute, it's an incredible resource filled with images tied to clinical and even genomic data. This context is what makes TCIA so powerful for serious research.
What I really appreciate about TCIA is how it organizes data into "Collections." These aren't just random piles of scans; they're curated datasets, often linked directly to a specific clinical trial or published paper. You know exactly what you're getting, from the patient cohort details to the imaging protocols used. You’re not just downloading a sample DICOM file; you’re accessing a piece of a larger scientific story.
My Advice: The sheer volume of data on TCIA can be overwhelming. Don't just dive in. Use the filters to narrow your search by modality (CT, MR, etc.) and body part right away. It will save you a ton of time.
OsiriX DICOM Image Library
Maybe you don't need a massive, research-grade dataset. Sometimes you just need a straightforward sample DICOM file to test a new feature or get a feel for the format. For that, the OsiriX DICOM Image Library is perfect.
Think of it less as a research archive and more as a developer's sandbox. It offers a wide variety of clean, simple examples—from a basic X-ray to a more complex 4D cardiac series. The beauty is in its simplicity. You can find what you need and download it in seconds without wrestling with a complex research portal.
Other Great Places to Look
TCIA and OsiriX are my usual first stops, but the community has built up a number of other valuable resources over the years. Depending on your specific project, one of these might have the exact data you're looking for.
- The Visible Human Project: This is a classic dataset from the U.S. National Library of Medicine. It contains complete, high-resolution CT and MRI scans of two cadavers (one male, one female). If you’re working on anatomical modeling or visualization software, this is a must-see.
- Medical Image Net: For the machine learning folks out there, this project is building a huge, curated medical image database inspired by the original ImageNet. It's designed specifically for training AI models.
- University & Hospital Archives: Don't forget to do a little digging on your own. Many academic medical centers put data from past research projects online. Searching for terms like "DICOM library" or "medical imaging data" along with a university's name can uncover some real gems.
Ultimately, the best repository depends entirely on what you're trying to accomplish. For deep, data-intensive research, nothing beats TCIA. But for quick tests and development work, the OsiriX library is far more efficient. Knowing the difference will get you the data you need, faster.
Generating Your Own Custom DICOM Files

Public repositories are great, but what happens when you need a sample DICOM file with a very specific, hard-to-find configuration? I’ve been there. Maybe you're testing how your software handles a rare imaging modality or a DICOM header with a funky private tag. In those moments, generating your own files is the only real solution.
Creating your own DICOM data gives you absolute control over every element, from the pixel data itself to the most obscure metadata tag. This is crucial for serious testing. You can build edge cases from scratch—think files with missing required tags or incorrectly formatted values—that you’d almost never see in the wild. This lets you proactively build a more robust application that won't choke on unexpected data in a live environment.
Let's walk through a couple of my favorite ways to get this done: a classic command-line toolkit and a really flexible Python library.
Using DCMTK for Command-Line Generation
If you're comfortable on the command line, the DICOM Toolkit (DCMTK) is an absolute must-have. It's a powerhouse open-source project with utilities for pretty much any DICOM task you can think of. For our purposes, the dcmgen utility is the star of the show, designed specifically for creating DICOM files from a simple configuration file.
The workflow is straightforward. You start by creating a text file where you define all the tags and values you want in your DICOM header. You can specify anything:
- Patient Information:
PatientName,PatientID,PatientBirthDate, etc. - Study Details:
StudyInstanceUID,StudyDate,AccessionNumber, and more. - Image Specifics:
Modality(like CT, MR, or XA) and theSOPClassUID.
With your configuration file saved, you just run a single command pointing dcmgen to it, and voilà—a fully compliant DICOM file is generated instantly. This approach is incredibly fast and perfect for scripting, letting you create dozens of file variations for automated testing with minimal effort.
Expert Tip: Never, ever reuse or manually create UIDs for tags like
StudyInstanceUIDorSOPInstanceUID. It’s a recipe for disaster in a real PACS environment. DCMTK has you covered with itsdcmuidtool, which generates new, compliant UIDs for you. Use it.
Creating DICOM Files with Python and Pydicom
When you need more dynamic control or want to embed DICOM generation directly into an application, the Python library Pydicom is the answer. It gives you a clean, object-oriented way to work with DICOM datasets, letting you build a file piece by piece in your code.
The real magic of Pydicom, in my opinion, is its ability to take a standard image like a PNG or JPEG and wrap it in a DICOM structure. This means you can take any image—say, a test phantom or a synthetic anomaly you've created—define the necessary metadata tags in your script, and merge them into a valid sample dicom file. It's incredibly useful for validating image processing algorithms.
The official Pydicom documentation has plenty of examples to get you up and running. By getting comfortable with both DCMTK and Pydicom, you'll have a complete toolkit to tackle any DICOM creation challenge your project throws at you.
How to Anonymize DICOM Files Correctly
When you're working with any sample DICOM file pulled from real-world data, you're handling sensitive information. A common—and frankly, dangerous—mistake is to just delete the patient's name from the header and call it a day. That’s like hiding your house key under the doormat. Real, effective anonymization goes much deeper to protect patient privacy and stay compliant with regulations like HIPAA.
Protected Health Information (PHI) is woven throughout the entire DICOM header. Sure, you have the obvious tags like PatientName (0010,0010) and PatientID (0010,0020), but sensitive data also hides in less obvious places. Things like dates, names of referring physicians, and hospital details can be pieced together to re-identify someone if you're not careful.
Beyond the Obvious Tags
The real trick is catching the data that doesn't scream "PHI" at first glance. Unique Identifiers (UIDs) are a classic example. Every study, series, and image gets its own unique ID generated by the scanner, stored in tags like StudyInstanceUID, SeriesInstanceUID, and SOPInstanceUID. While these long strings of numbers don't contain a name, they can often be traced right back to the original study in a hospital's PACS, completely undoing your anonymization work.
You also have to watch out for private tags. These are custom fields that equipment manufacturers add to the files. They might contain anything from proprietary scanner settings to, you guessed it, more patient information that standard anonymization profiles will miss entirely. Ignoring them leaves a potential backdoor to a patient's identity.
My Takeaway: Think of DICOM anonymization as a process of careful subtraction. You have to assume every single piece of metadata could potentially identify someone. The goal is to remove or replace anything and everything that could link a file back to a specific person, study, or institution.
This breakdown shows just how much metadata is packed into a typical DICOM file, separate from the actual image pixels.
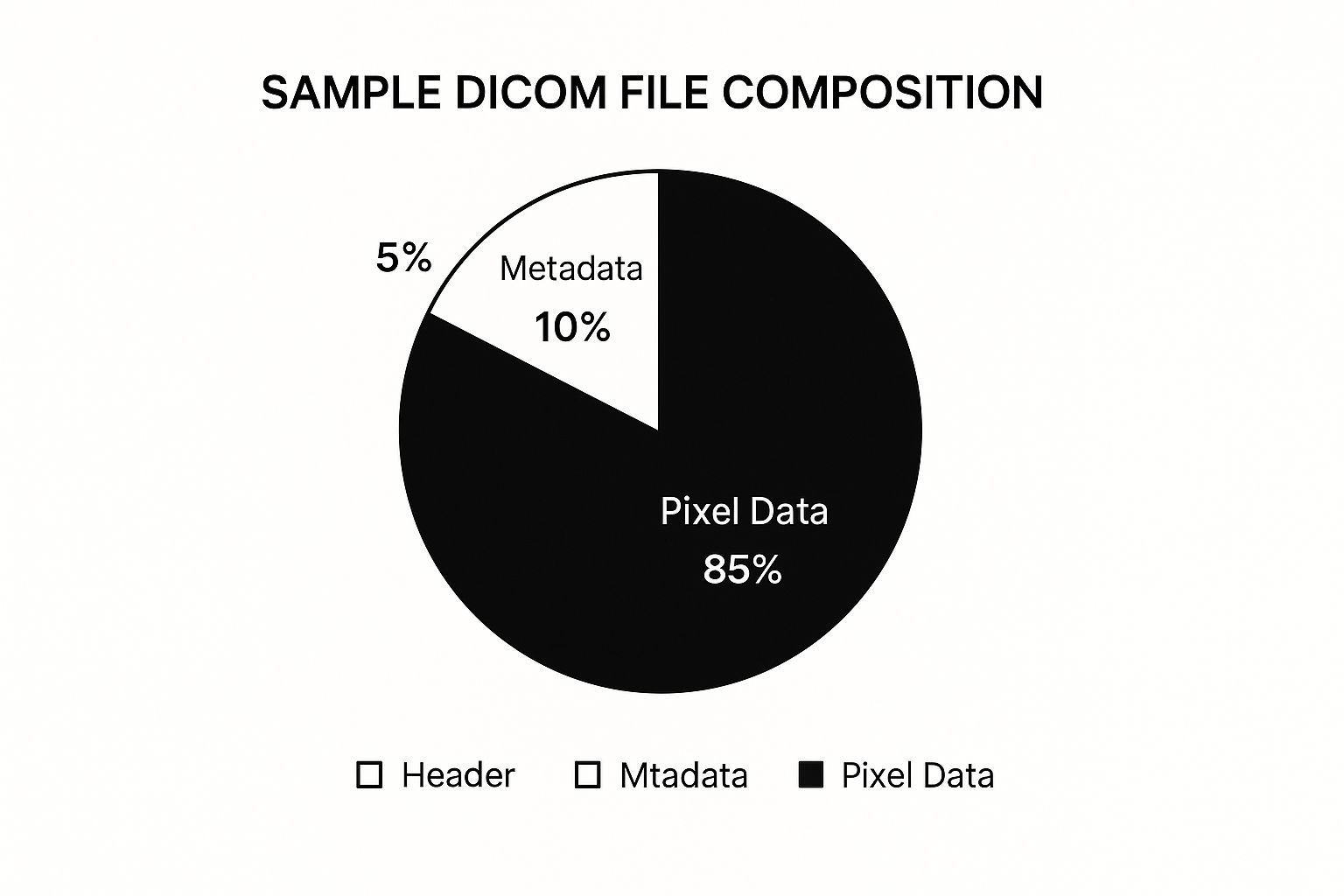
As you can see, while the pixel data is the biggest part of the file size, all the sensitive, identifying information lives in that smaller metadata section. That's where your focus needs to be.
Practical Anonymization Tools and Techniques
The good news is you don't have to go through every file and manually delete tags. There are some fantastic tools out there designed to handle this, each with its own approach.
To help you choose the right approach, here's a quick look at some common anonymization methods.
DICOM Anonymization Techniques Comparison
| Method/Tool | Ease of Use | Level of Control | Best For |
|---|---|---|---|
| Dedicated Tools | High (often GUI-based with presets) | Medium (relies on pre-configured profiles) | Quickly and safely processing large batches of files with standard needs. |
| Scripting (pydicom) | Low (requires coding knowledge) | High (total control over every tag and rule) | Complex, custom anonymization workflows and integration into other apps. |
| PACS Anonymizers | Medium (configured by an administrator) | High (powerful, rule-based policies) | Institutions needing to anonymize large data streams at the source. |
| Manual Editing | Very Low (tedious and error-prone) | Low (easy to miss critical tags) | Inspecting a single file, but not recommended for actual anonymization. |
Ultimately, choosing the right tool depends on your specific needs, whether you're a developer needing granular control or a researcher who just needs to clean a dataset quickly and reliably.
Your Anonymization Checklist
-
Dedicated Software: Tools built just for this, like DicomCleaner, are often the safest bet. They usually have user-friendly interfaces and come loaded with profiles based on the DICOM standard's official anonymization guidelines. They're perfect for processing batches of files with confidence.
-
Custom Scripts: If you’re a developer and need total control, Python's pydicom library is your best friend. You can write your own scripts to loop through every tag, apply custom logic, strip out all private tags, and even generate brand new, unlinked UIDs for every file.
The goal is to create a "clean" sample DICOM file—one that remains medically useful but is completely severed from its original source. While DICOM has its own specific challenges, brushing up on general data anonymization best practices provides a great foundation for protecting privacy across the board.
And my final piece of advice: always, always double-check your work. After running your anonymization process, open up a few files and inspect the headers yourself. It's the only way to be absolutely sure you didn't leave any sensitive data behind.
How to Inspect and View Your DICOM Files
So, you've got a sample DICOM file. Now what? The next logical step is to crack it open and see what’s inside. You can't just double-click and hope your standard image viewer will work; it won't. You need specialized software built to handle both the image itself and all the complex metadata packed into the header.
The good news is, there are some fantastic free tools out there that get the job done.
If you're on a Mac, Horos is the go-to open-source viewer. It's powerful and widely used. For Windows folks, the RadiAnt DICOM Viewer is a top choice, and its free trial is incredibly generous. Both of these let you do everything you'd expect: open single files, load entire patient studies, scroll through image stacks, and play with windowing and leveling to get the contrast just right.
Choosing the Right Viewer for Your Needs
Picking a viewer usually boils down to your operating system and what you’re trying to accomplish. Horos and RadiAnt are both excellent for just looking at images, but one might be a better fit depending on your specific project.
- Horos (macOS): This is a favorite among researchers and developers. Its real strength lies in its extensive plugin architecture and a very active community, which means you can often extend its capabilities to fit your unique needs.
- RadiAnt DICOM Viewer (Windows): I've always been impressed with how fast and lightweight RadiAnt is. It’s brilliant at loading and zipping through massive datasets without breaking a sweat, making it perfect for developers who are testing the performance of their own applications.
Here’s a look at the clean, intuitive interface of Horos. It’s a great example of a well-designed DICOM viewer.

You can see how it's designed with a clinical workflow in mind, letting you manage multiple studies on the left while digging into a specific image series in the main window.
Inspecting the Metadata Header
Just looking at the pixel data is only half the battle. The real gold is in the metadata. As a developer, this is where you'll spend a lot of time, verifying tag values and making sure your code is working as expected. It's also how you confirm that your anonymization process actually worked.
Thankfully, both Horos and RadiAnt have a built-in metadata inspector. This tool lays out every single tag in the file's header for you to examine.
A Quick Tip from Experience: When you're checking if a file is truly anonymized, don't just glance at the
PatientNametag and call it a day. I've seen protected information hiding in obscure places. Scroll through the entire tag list. Make sure all the UIDs have been properly replaced and that no hospital or doctor information is lingering in private or public tags.
For those of us who live in the command line, there's a much faster way. The DCMTK toolkit is an essential part of any DICOM developer's toolbox, and it includes a utility called dcmdump.
Just run dcmdump yourfile.dcm in your terminal, and it instantly spits out the entire header. It’s incredibly fast, easily scriptable, and perfect for automated testing or just doing a quick spot-check without firing up a full-blown GUI.
Common Questions About Sample DICOM Files
Diving into DICOM files for the first time can feel a little like learning a new language. You're bound to have questions, and certain issues pop up more often than others. Let's walk through some of the most frequent hurdles I see people encounter and how to clear them.
One of the first things people try is opening a DICOM file with their computer's default image viewer, only to be met with a "file not found" or "invalid format" error. This is completely normal. Remember, a DICOM file isn't a simple JPEG or PNG; it's a complex data object. You'll need a dedicated DICOM viewer like Horos or RadiAnt DICOM Viewer, or a programming library like Pydicom, to correctly interpret both the image and all the crucial metadata packed inside.
Another common point of confusion is the file extension. While .dcm is the standard, don't be surprised if you find files with no extension at all. This often happens when data is exported directly from a PACS system. If a file is giving you trouble, don't trust its name alone. A great pro-tip is to use a command-line tool like dcmdump to inspect its header. This will tell you instantly if you're dealing with a valid DICOM object.
Can a DICOM File Contain a Virus?
This is a smart question, especially when you're downloading files from public sources. While a DICOM file is fundamentally a data file, a specific design choice in the standard creates a potential vulnerability.
The standard allows for 128 arbitrary bytes at the very beginning of the file, right before the "DICM" prefix that identifies it. Malicious actors can exploit this space to hide executable code. This creates what's known as a polyglot file—one that looks like a perfectly normal medical image to a DICOM viewer but could execute malware if a user tries to run it as a program.
Always get your sample DICOM files from trusted, well-established repositories. This simple step is your best defense against accidentally downloading a compromised file.
Why Are Some DICOM Tags Unreadable?
Ever open up a file and see tags labeled something like "Private Tag (0029,10xx)"? These are private tags, which are custom fields added by equipment manufacturers to store their own proprietary information.
Because they aren't part of the official DICOM standard, most viewers won't know what they mean. Even worse, many anonymization tools can miss them entirely, leaving a backdoor for protected health information to leak through. It’s always good practice to scrub these thoroughly. For a broader look at how different industries structure their information, you can explore various sample technical document examples.
What’s the Difference Between a DICOM File and a DICOMDIR?
This one is pretty straightforward but important. Think of it this way:
- A DICOM File: This is a single file that usually holds one image instance. For example, one slice from a multi-slice CT scan.
- A DICOMDIR: This is an index file, not an image. It acts as a table of contents for a whole collection of DICOM files, like those you'd find on a CD from a patient study. It helps software quickly navigate the entire study without having to load and read every single file first.
Getting a handle on these key concepts will save you a lot of time and frustration as you work with sample DICOM data.
At PYCAD, we live and breathe the complexities of medical imaging data. From data anonymization to building and deploying sophisticated AI models, we handle it all. If you need expert help on your medical imaging project, visit us at PYCAD to see what we can do for you.






