Here’s the video version of this article, which may include some explanations that I forgot to include in the article.
Introduction
Regarding the difficulties that we can encounter when using traditional image processing tools, deep learning has emerged as the primary solution in the healthcare field.
Because medical images are more difficult to process than standard images (dense contrast, a wide range of variations in the human body…) Deep learning is used for classification, object detection, and especially segmentation tasks.
When it comes to segmentation, deep learning is used to segment human organs such as the liver, lung, and… or to segment tumors from different parts of the body.
There are many different types of medical images, such as MRI (which is mostly used for brain tumor segmentation), CT scans, PET scans, and so on.
This article will focus on CT scans, but the same operations can be applied to other types.
So we know that performing a deep learning task necessitates a number of steps, one of which is data preprocessing, which is the first thing we must do before launching a training. This is the topic of this article; we will discuss the tools available to us for performing this preprocess.
Preparing the data varies depending on the task; for example, classification is the easiest because we only need to prepare the images, whereas object detection and segmentation require us to prepare the image as well as the labels (bounding boxes or masks for segmentation).
In this article, we will use segmentation as an example, which can be used for tumor or organ segmentation.
In medical imaging, we can work with 2D images, which can be dicoms, JPGs, or PNGs, or 3D volumes, which are groups of slices, each slice is a 2D file (most of the time dicoms), and this group is a nifti file that represents the entire patient or only a portion of his body. Please read this article to understand the difference between dicom and nifti.
We will be working with 3D volumes in this article, so if you have 2D files that you want to convert into volumes, please see this article.
Tools we will use
To complete this task, we will use an open source framework called monai, which is based on PyTorch, which I used during my internship and found to be very useful.
Please see this link if you want to read its documentation.
Methodology
We will now begin coding our functions to perform the preprocessing for tumor segmentation in CT scans. The first step is to install the library using pip or conda, depending on which environment you’re using.
pip install monai
I strongly advise you to set up a virtual environment for your project because this library does not always work when installed directly in the system.
Then you need to install PyTorch and some dependencies of monai.
pip install torch
pip install torch-vision
pip install “monai-weekly[gdwon, nibabel,tqdm]”
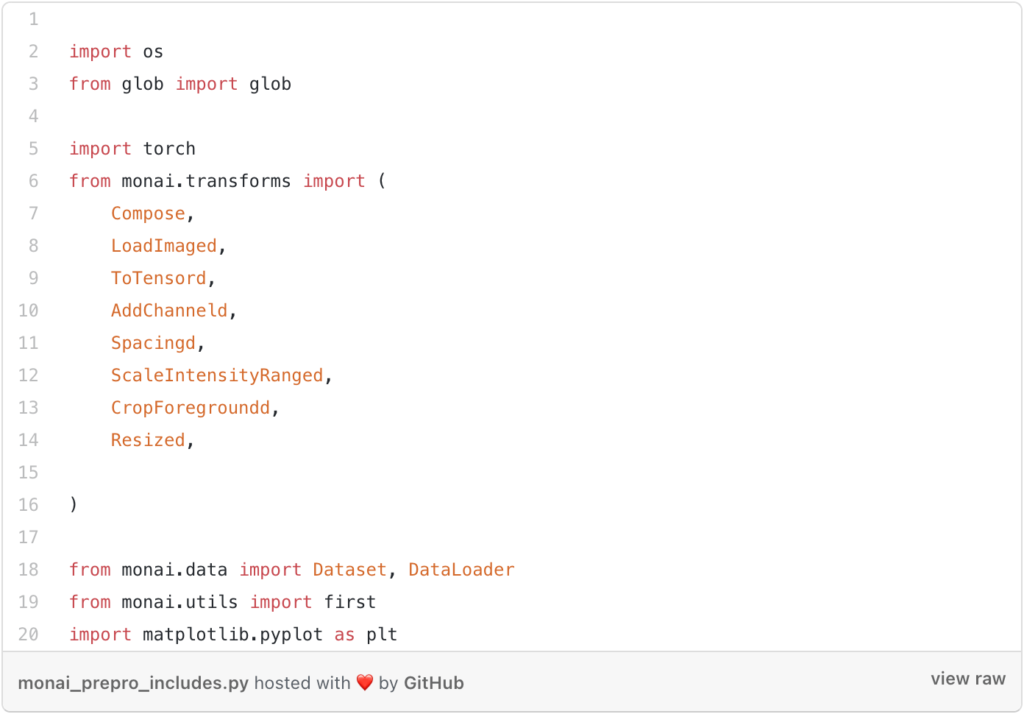
Then you need to include the libraries that you will need.
Now we’ll go over how to load data. There are two approaches to this.
1- The first is to load the images and masks individually (this is the way that you can use if you want to do image classification but it works also for segmentation).
2- The second method is to create a Python dictionary with two columns, one for the image paths and one for the label paths. The path of the image with the corresponding mask is then entered in each row.
Personally, I prefer the second method because, when we apply the transforms, we will be able to select just the key work of either the image or the label, or both, rather than creating transforms for the images and transforms for the labels (masks).
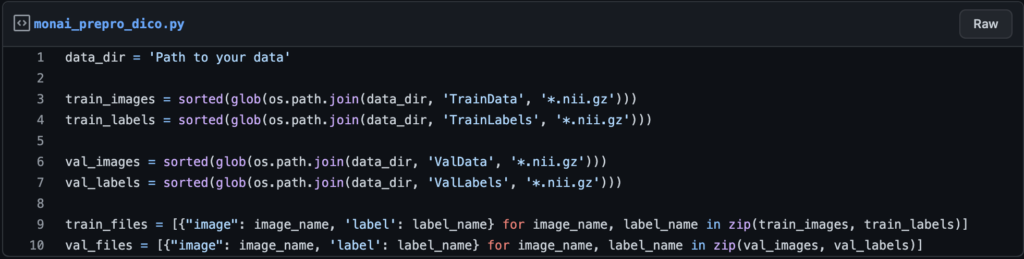
You must now create a variable that contains the path to the entire data set. In my case, I have four folders: TrainData, TrainLabels, and ValData, ValLabels for training images and masks, and ValData, ValLabels for validation images and masks.
So, to create a dictionary to store the paths, use the following lines of code:
After that, you have created the dictionary, which is simple if you are unfamiliar with it. If you want to refer to the first item in the dictionary, use index 0 as you would in a normal array, but the first item will have two columns, the first of which is the path to the image and the second of which is the path to the label (for more about this part, you can see the youtube video above).
The transforms
To apply multiple transforms to the same patient, we will use monai’s “compose” function, which allows you to combine any transformations you want (the ones defined in monai documentation).
There is something you should be aware of when applying any transform: some steps are required, while others are optional.
When using monai, the primary transforms are “Load image” to load the nifty files and “ToTensor” to convert the transformed data into torch tensors so that we can use it for training.
And now that we’ve covered the fundamental transforms, we’ll move on to the others. I’ll discuss five transforms that were particularly important during my internship.
AddChanneld: This function will add a channel to our image and label (when I say image, I mean the volume of multiple slices), because when we do tumor segmentation, we need a channel that plays the role of background or tumor.
Spacingd: This function will assist us in changing the voxel dimensions because we don’t know if a dataset of medical images was acquired with the same scan or with different scans, so they may have different voxel dimensions (width, height, depth), so we need to generalize all of them to the same dimensions.
ScalIntensityRanged: This function will assist us in performing two tasks at the same time: the first will be to change the contrast from that dense vision into something more visible, and the second will be to normalize the voxel values and place them between 0 and 1 so that the training will be faster.
CropForegroundd: This function will assist us in cropping out the empty regions of the image that we do not require, leaving only the region of interest.
Resized: Finally, this function is optional, but in my opinion, it is required if you used the cropforeground function, because the function that will do the crop will have the output with random dimensions depending on each patient, so if we do not add an operation to give the same dimensions to all patients, our model will not work.
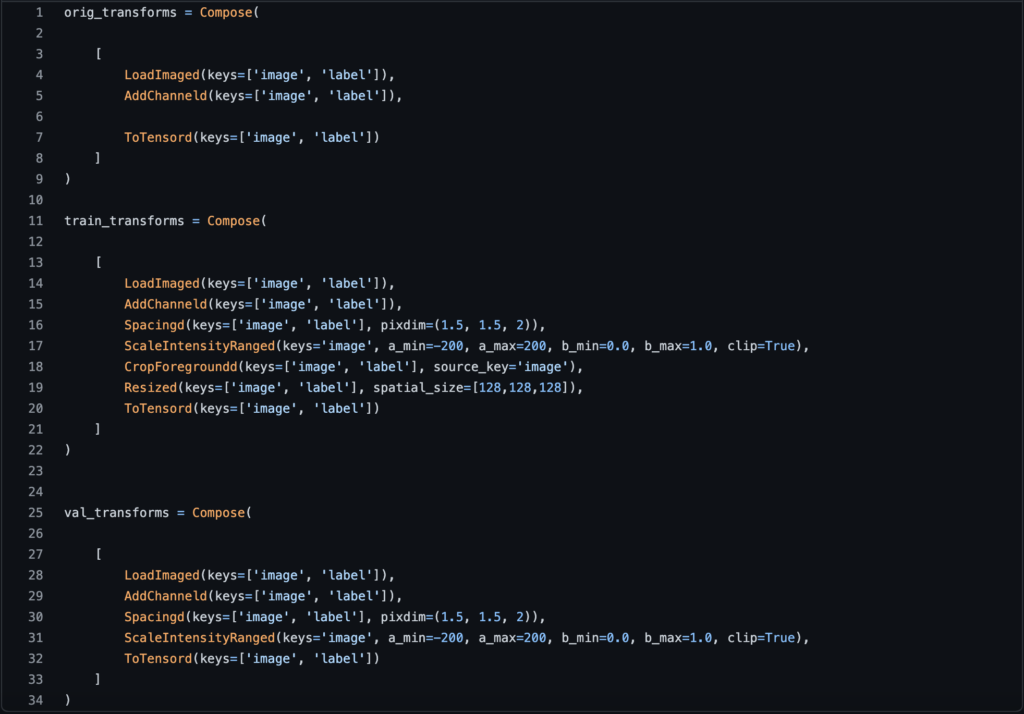
As you can see, there is a “d” at the end of each function we used, which is for the dictionary (if you are not using a dictionary then you need to remove it). And we added the parameter “keys” to each operation to specify whether we want to apply that transform to the images, the labels, or both. You can see that almost all of the time we applied the function to both the label and the image, but in the ScaleIntensityRange we only applied it to the image because we don’t need to change the intensity or normalize the labels’ values.
Data loader
Now, as with any deep learning code, we must load the data with its transform before we can begin training; to do so, two essential functions must be used.
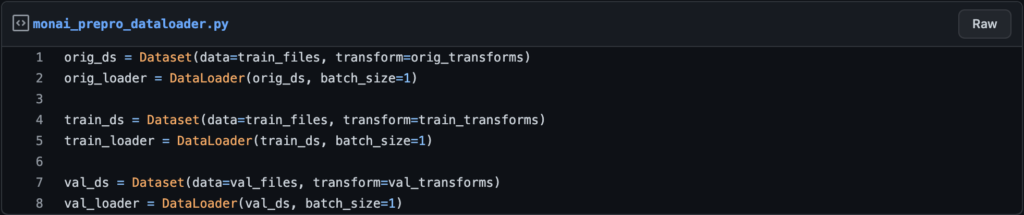
Dataset: This function will define the data along with its transforms, so if you have training and validation sets, you’ll need to create two “dataset” functions, one to combine the training data with its transforms and the other to combine the validation data with its transforms. Of course, if you apply the same transforms to the training and validation sets, you must use the same transforms in the parameter ‘transform’ in the function “dataset.”
Dataloader: This is the function that will load the data into RAM with a specific batch-size (it will create another channel to specify the index of the batch size during training) There will be two dataloaders, one for training and one for validation.
Plotting an example
After applying the transforms, you can plot some slices to see how the data differs with and without the transforms.
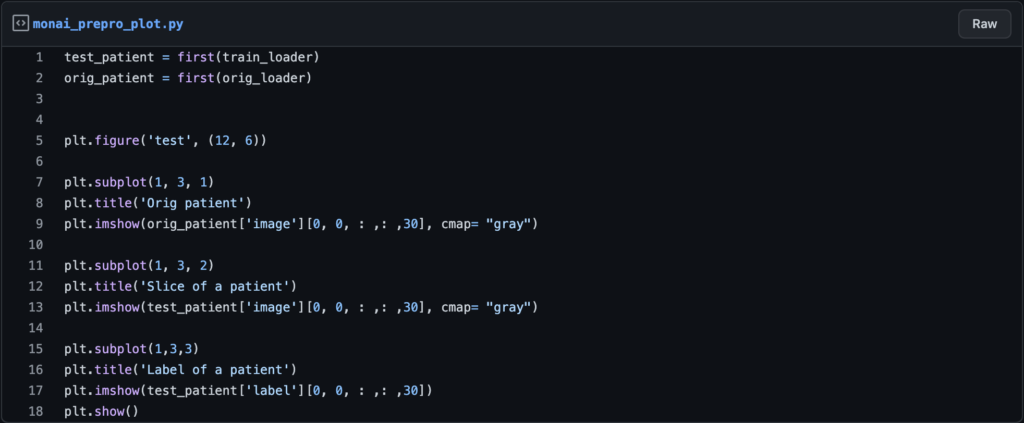
You can use the monai function “first” to get the first item from the dataloader, which will be the first patient.
Here are some lines of code to help you plot your examples.
Now let’s see the outputs with and without the transforms:
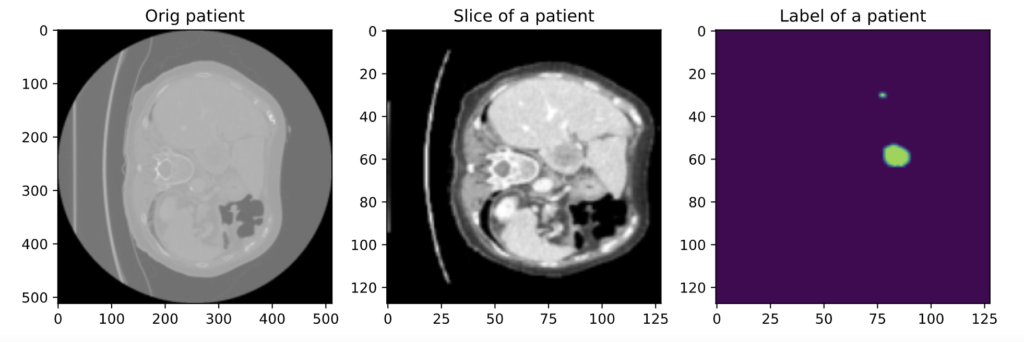
On the left is a slice with no transforms, on the middle is the same slice with the transforms we discussed, and on the right is the corresponding label.
So that’s how you use monai to prepare 3D volumes for segmentation. If you want to see more transformas, go to monai’s website. However, some transforms are designed for data augmentation rather than preprocessing. I’ll also write an article about it.
PS: I recommend watching the video because there are additional explanations that I may overlook in the article.
You can find the code from this link.