So, you've got a .mat file on your hands. Before we jump into how to open it, let's quickly cover what you're actually dealing with.
A MAT file is MATLAB's native format for saving your workspace. Think of it as a binary container that holds everything from variables and complex matrices to entire program functions.
What's Inside a .mat File?
If you're in fields like academic research, engineering, or data science, you’ll run into .mat files all the time. They aren't like simple CSVs; they’re more like structured packages that perfectly preserve the complex, multi-dimensional nature of scientific data. This makes them incredibly efficient for storing everything from sensor readings to simulation outputs in a compact, organized way.
This efficiency is exactly why they're so popular. When a researcher shares a dataset or a colleague sends over simulation results, it's often packaged as a .mat file. Knowing how to open and peek inside these files is a crucial skill, even if you don't live and breathe MATLAB. It's your key to accessing a massive amount of valuable data circulating in the scientific and engineering communities.
The file structure itself is well-defined. You'll mostly encounter two versions, Level 4 and Level 5, which are tied to older and newer MATLAB releases. Inside, data is stored in a binary format—not human-readable—created by MATLAB's save command and brought back to life with the load function. While you don't need to be an expert on the byte-by-byte layout, knowing it exists is helpful when you need to pull that data into another language.
For the technically curious, you can dig into the nitty-gritty details of the MAT-file format specifications.
Key Takeaway: A
.matfile is more than just raw data; it’s a complete snapshot of a computational workspace. It preserves the original variable names, data types, and complex structures, which is absolutely vital for reproducing scientific work.
Opening MAT Files Natively in MATLAB
If you have MATLAB, the most straightforward way to open a .mat file is just to double-click it. This dumps the file's entire contents right into your workspace. Simple, right?
Well, that approach works fine for small files, but it's a recipe for disaster with large datasets. I’ve seen it happen countless times—someone tries to open a massive file from a medical scan or a complex engineering simulation, and it just eats up all the system's RAM and crashes MATLAB. Not a great way to start your day.
A Smarter Way: The matfile Object
A much better approach, especially for those huge files, is to use the matfile object. This feature was a game-changer when it was introduced back in MATLAB R2011b. It was specifically designed to help you avoid that dreaded 'Out of Memory' error.
Essentially, it lets you peek inside a .mat file to see what’s there and then pull out only the specific pieces you need. You're not forced to load the whole thing into memory. You can get the full rundown on how it works from the official MathWorks documentation.
Putting matfile to Work with Partial Loading
This is where the real magic happens. Imagine you have a 10 GB .mat file from an MRI scan. All you need is the patient's metadata, which is just a tiny fraction of the total file size. Instead of loading gigabytes of image data you don't care about, you can use matfile to surgically extract only that small variable.
Here’s what that looks like in practice. It's clean and efficient.
% First, create a matfile object that links to your file
m = matfile('large_dataset.mat');
% You can see what variables are inside without loading them
whos('-file', 'large_dataset.mat');
% Now, load only the 'patientInfo' variable into your workspace
patientData = m.patientInfo;
Expert Tip: In any professional setting where you're dealing with serious data, using the
matfileobject is standard practice. It's a fundamental skill for making your code memory-efficient and robust. Honestly, it's non-negotiable if you want to avoid constant crashes.
The official documentation page gives a great overview of the function's syntax and purpose.

As the documentation shows, the object lets you access variables just like you'd access properties of an object (m.patientInfo). This syntax is what makes it so intuitive. By getting into this habit, you move from a brute-force loading method to a much more precise, memory-safe technique.
Accessing MAT Files Within the Python Ecosystem

If your workflow is centered on Python, you’re in luck. You don’t need a MATLAB license just to get at the data inside a .mat file. The Python scientific computing stack, specifically the SciPy library, is more than capable of handling these files. This is a game-changer for data scientists and researchers who live and breathe in Python's open-source world.
The magic happens in the scipy.io module. It has a function called loadmat that does all the heavy lifting. It reads the .mat file and translates its contents into a standard Python dictionary. The variable names from your MATLAB workspace become the keys, and the corresponding data arrays become the values. Simple.
This isn't some new or experimental feature, either. As open-source tools have matured, compatibility with proprietary formats is no longer the headache it used to be. You can find more details on the evolution of MAT file compatibility if you're curious about the history.
Loading Data with a Simple Snippet
Let's imagine you have a file called sensor_data.mat. Inside, you have two variables: time_vector and temp_readings. Loading this into Python is incredibly direct.
With just a couple of lines, you can pull that data right into a structure you already know how to work with.
from scipy.io import loadmat
Load the .mat file into a Python dictionary
mat_data = loadmat('sensor_data.mat')
Access the variables like any other dictionary item
time = mat_data['time_vector']
temperature = mat_data['temp_readings']
print(f"Loaded {len(time)} time points.")
Navigating the Loaded Data Structure
Once you load the data, you'll find everything is usually wrapped in NumPy arrays. But here's a heads-up: MATLAB's internal structures can sometimes create nested arrays that feel a bit odd in Python. For example, a single number like 5 in MATLAB might show up as a 2D array, [[5]], after being loaded.
Pro Tip: I almost always use the
squeeze_me=Trueargument when callingloadmat. This little flag tells SciPy to remove any extra, single-dimension entries from your arrays. It cleans up the data structure beautifully, making it much easier to handle right out of the gate.
This parameter saves you from writing boilerplate code just to flatten or reshape arrays. It’s a small tweak that seriously improves the readability of your scripts, especially when you're just trying to open a MAT file for a quick analysis or plot.
Using Open Source Alternatives Like GNU Octave

If you need the number-crunching power of MATLAB but don't have a commercial license, GNU Octave is your best friend. It’s a powerful, free, and open-source tool that’s been my go-to for years when I’m working on personal projects or collaborating with academics.
What makes it so great is its high compatibility with MATLAB syntax. This means you can often run MATLAB code with few, if any, changes. And yes, it handles .mat files natively, which is exactly what we need.
The process for loading data will feel second nature if you're coming from MATLAB. The commands are practically identical, so there's almost no learning curve. You just load the .mat file, and its variables appear in your Octave workspace, ready to go.
Loading and Inspecting Data in Octave
To get started, you'll use the familiar load command. Let's imagine you have a file called experiment_data.mat.
The syntax is just as clean and simple as in MATLAB, making it incredibly easy to switch between the two.
% Load the entire contents of the .mat file into the workspace
load('experiment_data.mat');
% Display a list of variables currently in the workspace
whos
% Access a specific variable by its name
disp(my_matrix);
Once you run load, all the variables from the file are dumped right into your workspace. I always follow it up with the whos command—it gives you a quick, clean summary of everything you just imported, including variable names, dimensions, and how much memory they're taking up.
A Quick Heads-Up: While Octave is an amazing tool, it's not a perfect carbon copy of MATLAB. For really specialized tasks that rely on specific toolboxes or newer object-oriented programming features, you might hit a few snags. But for core data handling and matrix math—including opening
.matfiles—it’s an exceptionally reliable and free alternative.
Choosing the Right Tool for Opening MAT Files
So, you've got a .mat file and need to get inside. What's the best way to do it? Honestly, there’s no single right answer—it really depends on your setup and what you're trying to accomplish.
Are you working in a Python-first environment? Do you have a MATLAB license? Is this a one-off task or part of a daily workflow? The best tool for the job comes down to your budget, your go-to programming language, and the sheer size of the data you’re wrangling.
Comparing Your Options
Let's break down the main contenders: MATLAB itself, Python with the SciPy library, and the open-source alternative, GNU Octave. Each has its own strengths and weaknesses.
- Cost: This is often the biggest deciding factor. MATLAB is a commercial product and requires a paid license. Python and Octave, on the other hand, are completely free.
- Performance: For truly massive files (we're talking gigabytes), MATLAB's native
matfileobject is tough to beat. That said, Python'sh5pylibrary does an excellent job with v7.3 MAT files, which are based on the HDF5 format. - Ease of Use: If you're just starting out, Octave is incredibly approachable. It’s designed to be a free alternative to MATLAB, so the syntax and environment will feel very familiar to anyone coming from that world.
- Compatibility: This is where MATLAB shines. It guarantees 100% compatibility with every data type and complex object you might find in a .mat file. Python can sometimes struggle to correctly interpret more obscure or custom MATLAB structures.
This chart gives you a sense of how performance can differ when working with .mat files.
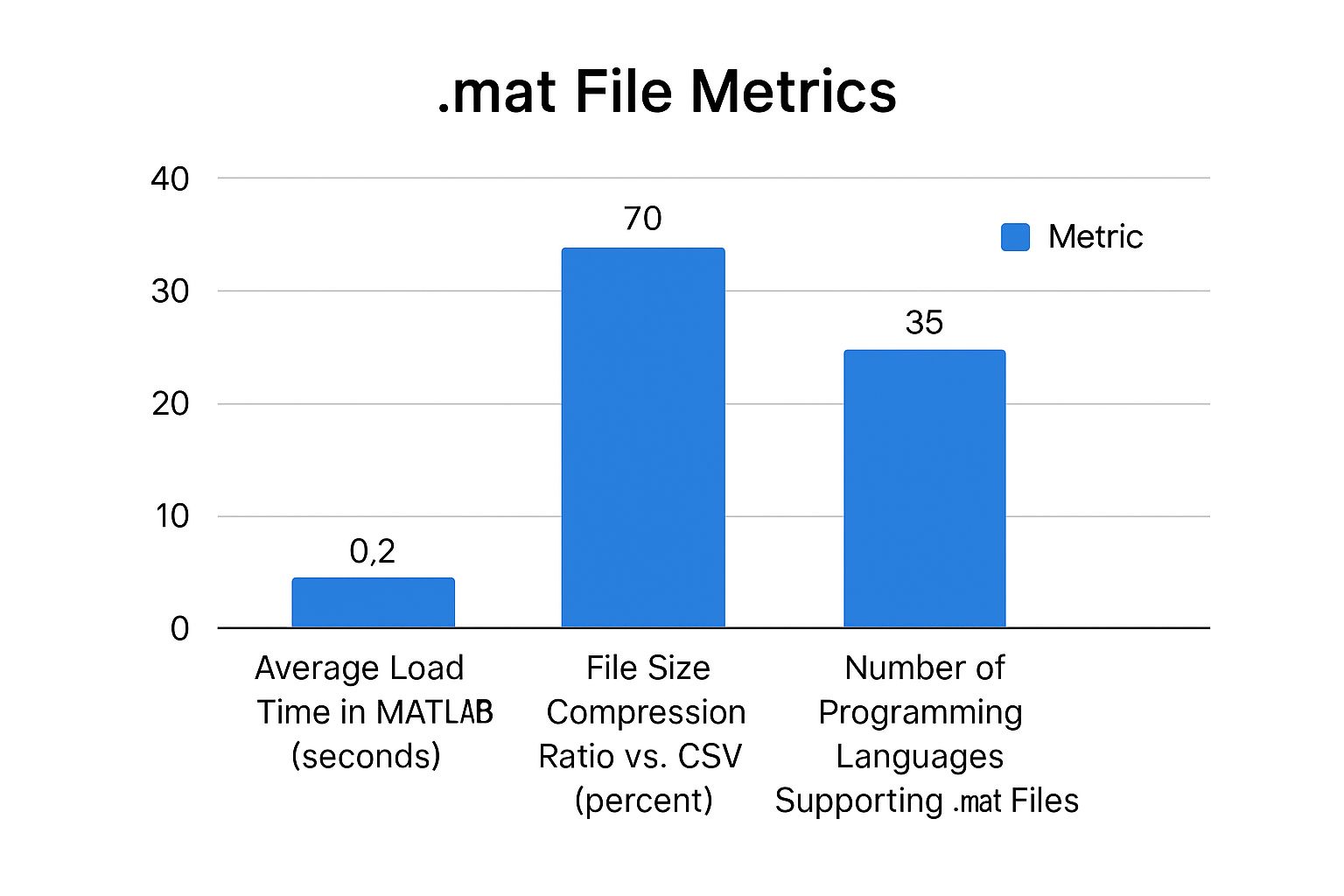
As you can see, even though MAT files have great compression, the time it takes to load them can change quite a bit depending on your toolchain.
From my own experience, Python or Octave are the obvious choices in academic or research settings where every penny counts. But if you're in a corporate environment where rock-solid compatibility and peak performance are non-negotiable, sticking with the native MATLAB environment is almost always worth the investment.
To make the decision even clearer, here’s a quick side-by-side comparison.
MAT File Access Method Comparison
| Method | Primary Use Case | Cost | Large File Performance | Ease of Use |
|---|---|---|---|---|
| MATLAB | Professional engineering, native data analysis, toolbox-heavy workflows | Paid License | Excellent | High (native environment) |
| Python (SciPy/h5py) | Integrating MATLAB data into Python-based ML/AI or data science pipelines | Free | Good to Excellent | Moderate (requires libraries) |
| GNU Octave | Academic use, prototyping, free alternative to MATLAB for basic tasks | Free | Fair | High (MATLAB clone) |
| HDF5 Viewers | Quick inspection and verification of v7.3 file contents without coding | Free | Good | Very High (GUI-based) |
Ultimately, the best tool is the one that fits seamlessly into your existing workflow without causing unnecessary headaches. For most, Python strikes a great balance, but it's good to know you have options.
Common Questions (and Fixes) When Working With MAT Files
Even with the right tools, MAT files can throw a few curveballs your way. If you've ever opened one and thought, "Wait, why does my data look like that?", you're not alone. Let's walk through some of the most common hangups people run into.
Why Is My Data So Nested in Python?
One of the first things you’ll notice when you use scipy.io.loadmat is that your data comes back as a Python dictionary. This is actually a good thing! It means the original variable names from your MATLAB session are preserved as the dictionary keys, which keeps things organized.
But then you see the values. Even a single number might look like [[5]]. This is because, in MATLAB's world, everything is a matrix. A scalar is just a 1×1 matrix. To flatten this out and make your data easier to work with, just add the squeeze_me=True argument when you call loadmat. It’s a simple trick that gets rid of those extra, unnecessary dimensions.
What’s the Deal With Newer v7.3 MAT Files?
Yes, you can absolutely open the newer v7.3 MAT files, but there's a little secret to it. These modern MAT files are actually HDF5 files in disguise. The good news is that scipy.io.loadmat is smart enough to handle them, but it needs a helper library to do it: h5py.
If you try to open a v7.3 file and get an error, it's almost always because
h5pyis missing. Just runpip install h5pyin your terminal, and you'll be all set to handle any MAT file, old or new.
What's the Best Free Way to Just Peek Inside a MAT File?
If you don't want to write any code and just need to quickly see what's inside a MAT file, GNU Octave is your best bet. It has a graphical interface and a variable editor that will feel instantly familiar if you've ever used MATLAB. You can just load the file and browse through the variables.
For a more programmatic approach, Python with SciPy and h5py is a fantastic, powerful, and totally free combination for digging into the contents of any MAT file you come across.
At PYCAD, we spend our days wrangling complex medical imaging data, including the very formats you see in research. If you're building AI for medical diagnostics and need to accelerate your project, see how our expertise in data management and model deployment can help at https://pycad.co.






