Picture this: you have an entire library's worth of complex scientific information—satellite images, genomic sequences, climate models—and you need to store it all in a single, well-organized, and easily searchable place. This is precisely the challenge that the HDF file format was designed to solve. It acts as a specialized container for massive, complex datasets that would completely overwhelm simpler formats like CSV.
Why the HDF File Format Is Essential for Big Data
When you're dealing with big data, you quickly realize that not all file formats are created equal. A simple text file might be fine for a grocery list, but it's utterly useless for handling terabytes of multi-dimensional data from a supercomputer simulation or a high-resolution MRI scan. This is where the Hierarchical Data Format, or HDF, truly shines.
It helps to think of an HDF file not as a single flat document, but as a sophisticated digital filing cabinet. Inside this cabinet, you can create a structured hierarchy of folders (which HDF calls Groups) and place your actual data arrays (called Datasets) within them. This built-in organization is fundamental for taming complexity.
A Self-Describing Data Container
One of the most powerful aspects of the HDF file format is that it’s self-describing. This means the metadata—the crucial "data about the data"—is stored right alongside the information it describes. You can attach all kinds of context directly to your datasets, such as:
- Measurement units (e.g., Celsius, meters per second)
- Timestamps for data collection
- Sensor calibration settings or equipment models
- Notes from the researcher or specific experiment parameters
Bundling the data and its descriptive metadata into a single, portable file is a game-changer. It prevents critical context from getting lost or misinterpreted over time. Someone can open that file years later and understand exactly what the data means without needing a separate instruction manual. To make the most of this powerful feature, following essential data quality best practices ensures the stored information remains accurate and reliable.
To really grasp what makes HDF so effective, let's break down its core attributes.
HDF File Format Key Features at a Glance
The table below summarizes the features that have made HDF a go-to standard for scientific and large-scale data applications.
| Feature | Description | Primary Benefit |
|---|---|---|
| Hierarchical Structure | Organizes data into a tree-like system of groups and datasets, similar to a computer's file system. | Manages complex, heterogeneous data in a single, organized file. |
| Self-Describing | Allows metadata to be stored directly alongside the data it describes, within the same file. | Ensures data portability and long-term usability; context is never lost. |
| Support for Large Datasets | Designed to handle datasets that can reach terabytes or even petabytes in size. | Accommodates the scale of modern scientific instruments and simulations. |
| Parallel I/O | Enables multiple processes to read from or write to a single HDF file simultaneously. | Drastically speeds up data access and processing in high-performance computing environments. |
| Data Compression | Includes built-in support for various compression algorithms (like Gzip) to reduce file size. | Saves storage space and reduces data transfer times without sacrificing integrity. |
These features combine to create a robust and highly efficient format for modern data challenges.
HDF5 was specifically engineered to overcome the file size and performance limitations of older data formats. It supports virtually unlimited file sizes and includes advanced capabilities like parallel I/O, which is absolutely critical for high-performance computing tasks where speed is paramount.
In the end, it’s this unique ability to store, organize, and manage massive and complex information efficiently that makes the HDF file format a cornerstone of modern research and data analysis across countless fields.
From University Project to Global Standard: The Story of HDF

Every great technology has an origin story. For the HDF file format, that story doesn't start in a corporate R&D lab but in the world of academic and government research, born from a pressing need to manage scientific data on an unprecedented scale.
Our story begins back in 1987 at the National Center for Supercomputing Applications (NCSA). Researchers there were dealing with the incredibly complex outputs from their supercomputers and needed a better way to handle it all. Their solution was the very first version of HDF, which quickly caught on for its clever ability to pack different kinds of data into one organized file.
NASA’s Stamp of Approval and the Rise of HDF4
The real turning point came in the early 1990s. NASA was gearing up for its massive Earth Observing System (EOS) project and needed a standard format to handle the flood of satellite data they were about to collect. The format had to be tough, reliable, and able to grow with the project.
After a grueling two-year evaluation of 15 different formats, NASA chose HDF as its official standard. This decision was a massive vote of confidence that cemented HDF’s place in the scientific community. You can dive deeper into the history of HDF and its early days to see just how foundational this adoption was.
This collaboration led to the formalization of what we now call HDF4. For its time, HDF4 was a huge leap forward, offering a flexible way to store datasets alongside their all-important metadata. But as computing power exploded and datasets ballooned, its seams began to show.
The biggest headaches with HDF4 were its hard 2-gigabyte file size limit and a somewhat clunky data model. These technical walls made it a real struggle to work with the enormous datasets coming from modern scientific instruments and simulations.
Faced with these growing pains, the developers at NCSA knew a simple patch wouldn't cut it. They went back to the drawing board, not to fix HDF4, but to build something entirely new for the next era of scientific computing.
A New Beginning: HDF5 and The HDF Group
This massive effort gave us HDF5, a complete redesign that blew past the old limitations. HDF5 brought a much more versatile data model and, most importantly, completely eliminated the file size cap. It was built to scale for just about any data challenge you could throw at it. It also came packed with features essential for high-performance computing, like parallel I/O.
To safeguard the format’s future as an open standard, The HDF Group was founded in 2008. This non-profit organization became the official steward of HDF, offering support and steering its development for a worldwide community of users. Today, HDF5 is the go-to standard for new projects, a perfect example of how a collaborative, forward-thinking idea can grow from a university project into a global powerhouse.
A Look Inside the HDF5 File Structure
To really get why HDF is so useful, you have to peek under the hood. The best way to think about an HDF5 file isn’t as a single, flat file, but as a mini, self-contained file system. It’s like a super-smart zip archive built specifically for complex data.
This internal structure is what makes HDF5 so organized and efficient. It's all built from just a few core components that work together beautifully. Once you get these, you understand HDF5.
Groups: The Folders of HDF5
The primary way you organize things in an HDF5 file is with a Group. A Group is almost exactly like a folder or directory on your computer. Its job is to hold other things—either more Groups or Datasets—letting you build a logical, nested structure for your data.
For example, you might create a top-level Group for a research experiment. Inside that, you could have subgroups for raw sensor data, processed results, and simulation parameters. This keeps everything tidy and makes the file easy for anyone to understand at a glance.
Datasets: Where the Data Lives
If Groups are the folders, then Datasets are the files inside them. A Dataset is a multidimensional array that holds your actual data. This could be a 2D medical image, a 3D volume from a CT scan, or a simple 1D array of temperature readings over time.
This is where the numbers are stored. Each Dataset contains a uniform chunk of data, much like a NumPy array, and it's the final container for your measurements or outputs. The interplay between Groups and Datasets is what lets you arrange incredibly complex data in a way that still makes perfect sense.
This diagram shows just how simple and powerful this hierarchy is, with Groups acting as containers for Datasets.
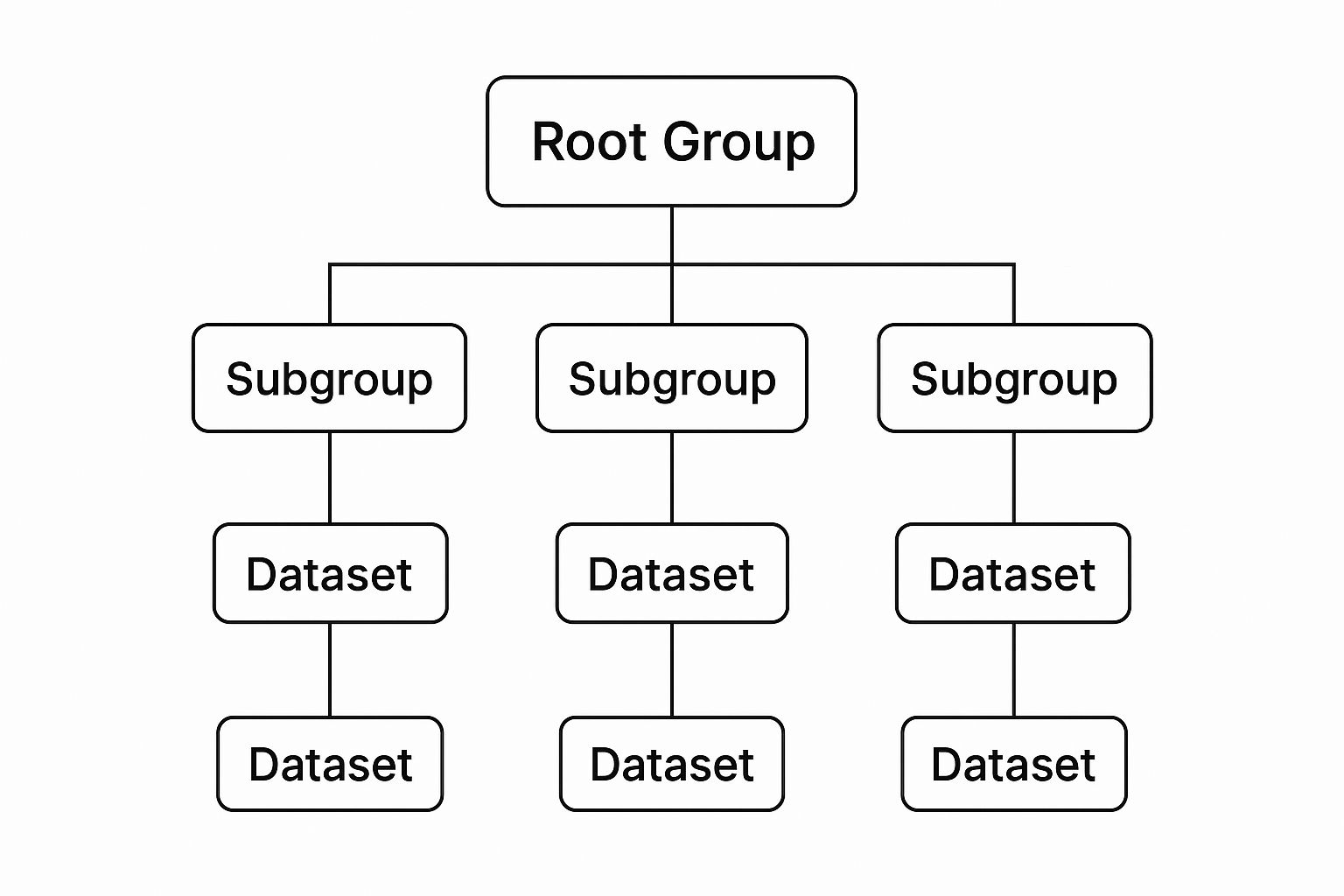
As you can see, a top-level "Root Group" can branch out into Subgroups and Datasets, creating a tree-like structure that’s easy to navigate and expand.
Attributes: The Sticky Notes for Your Data
The final piece of the puzzle is Attributes. I like to think of them as digital sticky notes you can attach to any Group or Dataset. They’re small, named pieces of metadata meant to add context right where you need it.
You might use an Attribute to store the units of a Dataset (like "Celsius" or "mm"), the date an experiment was conducted, or the version of the software used for processing. It’s this self-describing quality that really sets HDF apart.
Together, these three components—Groups, Datasets, and Attributes—create a single file that is portable, organized, and self-documenting. This elegant design is precisely why HDF is so trusted for handling the messy, high-volume data found in science and medical imaging.
Comparing the HDF4 and HDF5 Formats

While they share the same family name, it's a common mistake to think of HDF4 and HDF5 as just different versions of the same thing. They aren't. HDF5 was a complete, from-the-ground-up redesign to fix the deep-seated limitations that were holding HDF4 back as data sets grew exponentially.
Think of it this way: comparing HDF4 to HDF5 is like comparing a horse-drawn carriage to a modern electric car. Sure, both are technically forms of transportation, but they’re built on entirely different engineering principles for completely different eras. You might still find old HDF4 files in legacy scientific archives, but for any new work, HDF5 is the only way to go.
Key Architectural Differences
The biggest deal-breaker for HDF4 was its hard 2 GB file size limit. In an era of terabyte-scale datasets from medical imaging, climate modeling, and particle physics, this limitation simply made it obsolete.
HDF5, on the other hand, was built for the future with virtually unlimited file sizes. It also introduced a much more sophisticated and flexible data model. HDF4 was stuck with a few rigid, predefined data types, whereas HDF5 lets you define your own, allowing for far more complex data structures.
HDF5's redesign wasn't just about making files bigger; it was about performance. It brought native support for critical features like parallel I/O, which allows multiple processes to read and write to the same file at once—an absolute must for high-performance computing.
The HDF5 format we use today emerged in the late 1990s, specifically to solve the scalability and usability problems of HDF4. It wasn't an update; it was a new format with a new API that supported modern needs like chunked storage, compression, and better metadata handling. Since 2008, development has been managed by The HDF Group. You can dive deeper into the history of HDF5's evolution if you're curious about the technical journey.
A Side-by-Side Comparison
To really see the difference, a direct comparison helps. The table below breaks down why these two formats are worlds apart.
HDF4 vs HDF5 Key Differences
| Feature | HDF4 | HDF5 |
|---|---|---|
| Max File Size | 2 GB | Practically unlimited |
| Data Model | Limited, predefined data types | Flexible, user-defined data types |
| API | Multiple APIs for different data models | A single, unified API |
| Parallel I/O | Not supported | Fully supported |
| Threading | Not thread-safe | Thread-safe |
| Objects per File | Limited to 20,000 | Practically unlimited |
This table makes it clear why HDF5 wasn't just an upgrade—it was a necessary reinvention. For any modern project dealing with serious data, HDF5 is the undisputed standard. It delivers the scalability and performance that today’s demanding applications need.
How to Read and Write HDF5 Files with Python

Alright, enough with the theory—let's get our hands dirty. For anyone working in Python, the go-to library for handling HDF5 is h5py. It’s a beautifully designed package that makes interacting with HDF5 files feel natural and, well, Pythonic. Groups behave like dictionaries, and datasets act a lot like NumPy arrays.
We’re going to walk through the entire process from scratch: creating a file, building a clean data structure inside it, and then reading everything back out. By the time we're done, you'll see just how straightforward it is.
First thing's first, make sure you have h5py installed. A quick pip install h5py in your terminal will do the trick.
Creating Your First HDF5 File
Opening an HDF5 file with h5py feels a lot like opening any other file in Python. We use a with statement, which is great practice because it automatically handles closing the file for us, preventing any nasty data corruption issues.
import h5py
import numpy as np
Create a new HDF5 file in write mode ('w')
with h5py.File('my_first_hdf_file.h5', 'w') as f:
print("File created successfully!")
The 'w' mode tells h5py to create a new file. Just be careful, as it will overwrite any existing file with the same name. You can also use 'r' for read-only access or 'a' to append (read and write) to a file without wiping it clean.
Organizing Data with Groups and Datasets
With our file open, we can start building its internal hierarchy. Let's imagine we're storing data from an experiment. A logical first step is to create a group to hold everything related to that experiment. Then, inside that group, we can save our actual data—a NumPy array—as a dataset.
This is where the "folder and file" analogy really clicks into place.
Re-open the file in append mode ('a') to add data
with h5py.File('my_first_hdf_file.h5', 'a') as f:
# Create a group to organize our data
experiment_group = f.create_group('experiment_data')
# Generate some sample data
sensor_readings = np.random.rand(100, 3) # 100 readings, 3 sensors
# Create a dataset within the group to store the data
dset = experiment_group.create_dataset('sensor_readings', data=sensor_readings)
print("Group and dataset created.")
Think about what we just did: experiment_data is our new directory, and sensor_readings is the file inside it that holds our array. Simple and organized.
Adding Context with Attributes
One of the most powerful features of the HDF file format is that it's self-describing. We can attach metadata—called attributes—directly to our groups and datasets. This is how you give your data context.
Think of attributes as digital sticky notes. They help you—and anyone else who uses your file—understand the data's origin, units, and purpose without digging through separate documentation. This habit is absolutely essential for creating shareable and reproducible scientific data.
Let's add a few helpful attributes to the dataset we just created.
with h5py.File('my_first_hdf_file.h5', 'a') as f:
# Access the dataset we created earlier
dset = f['/experiment_data/sensor_readings']
# Add attributes to the dataset
dset.attrs['units'] = 'Volts'
dset.attrs['sensor_model'] = 'Model-X42'
dset.attrs['timestamp'] = '2024-10-26T10:00:00Z'
print("Attributes added.")
Now, anyone opening this file immediately knows the measurements are in Volts and which sensor they came from.
Reading Data from an HDF5 File
Finally, let's pull our data back out. Reading from an HDF5 file with h5py is incredibly intuitive; you access groups and datasets just like you would with Python dictionaries and their keys.
with h5py.File('my_first_hdf_file.h5', 'r') as f:
# Access the dataset using its full path
data = f['/experiment_data/sensor_readings'][:]
# Read an attribute
units = f['/experiment_data/sensor_readings'].attrs['units']
print(f"Dataset shape: {data.shape}")
print(f"Data units: {units}")
That little [:] at the end is important. It’s what tells h5py to load the entire dataset from the disk into a NumPy array in your computer's memory. And just like that, you've completed the full cycle: create, write, annotate, and read an HDF5 file.
Answering Your HDF Questions
Whenever you’re getting your hands dirty with a new file format, questions are bound to come up. With HDF, and especially HDF5, I see the same handful of queries pop up time and time again. Let's walk through them, because getting these answers straight will make it crystal clear when and why HDF5 is often the best tool for the job.
Think of this as a quick-start guide to help you sidestep common hurdles and feel more confident in your data strategy.
When Should I Use HDF5 Instead of CSV or JSON?
This is probably the most common question, and the answer really boils down to the shape and size of your data. While CSV and JSON are fantastic for many things, they have breaking points that HDF5 was built to handle gracefully.
You’ll want to reach for HDF5 when you're dealing with:
- Large, multi-dimensional numerical data. This is HDF5’s sweet spot. We're talking about massive datasets from scientific simulations, 3D medical scans like CT or MRI, sprawling satellite images, or high-frequency sensor logs.
- Performance-critical applications. Because HDF5 is a binary format, it supports partial I/O. This means you can grab a single slice from a massive 3D array without having to load the entire multi-gigabyte file into memory first. It's a game-changer.
- Complex, mixed datasets. Imagine needing to store images, data tables, and configuration notes all in one place. HDF5’s group structure is perfect for keeping all that related-but-different data neatly organized within a single file.
CSV is great for simple, flat tables, and JSON is king for nested text and configs. But once you start throwing large numerical arrays at them, they get incredibly slow and hog memory. For scientific and medical work, HDF5 is almost always the right choice.
What Are the Biggest Advantages of HDF5?
The real magic of the HDF file format comes down to three core strengths. These are the pillars that have made it a go-to standard in some of the most data-intensive fields on the planet.
Here’s what makes it so powerful:
- Portability: Everything—the raw data and all its descriptive metadata—is bundled into one self-contained, self-describing file. This makes sharing it with a colleague or archiving it for 10 years incredibly simple. The context never gets lost.
- Performance: Its binary structure, combined with features like chunking and compression, allows for lightning-fast access to just the piece of data you need. It also supports parallel I/O, which is a must-have in high-performance computing.
- Scalability: For all practical purposes, there are no limits on the size of an HDF5 file or how many objects you can stuff inside it. This future-proofs your work, ensuring the format won't become a bottleneck as your datasets inevitably grow.
These features work together to create a rock-solid system for managing data that's both enormous in scale and intricate in structure. It’s an architecture built to last, ensuring your data remains accessible and understandable for years.
What Tools Can I Use to View HDF5 Files?
Good news—you don't always have to write code just to peek inside an HDF5 file. There’s a whole ecosystem of user-friendly tools that give you a graphical window into your data, letting you explore its structure without touching a single line of Python or C++.
For a quick visual look, the most popular option by far is HDFView. It's a free, cross-platform tool from The HDF Group that gives you a simple tree view of your groups and datasets. You can just click through the hierarchy, view the data arrays, and check out the metadata.
Beyond that, many scientific environments like MATLAB and visualization tools like ParaView and Panoply have built-in support for the HDF file format, since it’s so common in research. And for those who live in the terminal, the standard HDF5 software comes with handy command-line utilities like h5dump and h5ls for quick inspections.
At PYCAD, we use powerful data formats like HDF5 every day to build and deploy advanced AI solutions for medical imaging. If you're looking to bring more accuracy and efficiency to your diagnostic workflows, see how we can help.






