High-quality data is the lifeblood of innovation in medical AI, yet finding the right medical image dataset can be a monumental challenge for researchers and developers. These specialized collections are crucial for training, testing, and validating algorithms that can revolutionize diagnostics, from detecting cancers earlier to automating complex organ segmentation. However, navigating the landscape of public and private repositories, each with its own access requirements, data formats, and annotation quality, requires a clear roadmap. This guide cuts through the complexity by providing a detailed breakdown of 12 essential platforms and repositories.
We’ll explore their specific modalities (like X-ray, MRI, and CT), ideal use cases, and honest limitations to help you select the perfect resource to fuel your next breakthrough. Beyond simply identifying datasets, leveraging them effectively requires understanding and implementing robust data handling. For a deeper dive into this area, we recommend this practical guide to research data management.
Our goal is straightforward: to help you quickly identify the most suitable medical image dataset for your specific project, whether you’re developing a new diagnostic tool or conducting academic research. Each entry in our list includes direct links and key details, saving you valuable time and effort in your search.
Btw, we build web DICOM viewers and custom medical imaging CRMs for our clients. Check out our portfolio.
1. The Cancer Imaging Archive (TCIA)
The Cancer Imaging Archive (TCIA) is an indispensable, publicly funded resource for anyone working in oncology AI. It serves as a large-scale repository of de-identified medical images, primarily focused on cancer, and is an essential starting point for training and validating diagnostic algorithms. What sets TCIA apart is its rich, multi-modal data integration; images are often linked with corresponding clinical outcomes, genomic data, and pathology reports. This provides a holistic view crucial for developing sophisticated predictive models.

Key Features and Use Cases
The platform is more than just a data dump; it’s a well-structured ecosystem for reproducible research. The data is standardized in DICOM format, ensuring interoperability.
- Ideal Use Case: Excellent for training computer-aided detection (CADe) and diagnosis (CADx) systems for various cancers, such as lung, breast, and brain tumors. The associated clinical data supports projects aiming to predict patient prognosis or treatment response from imaging features.
- Access Requirements: While the majority of datasets are freely and publicly accessible, some collections are restricted and require an application to protect patient privacy or due to specific use agreements.
- Practical Tip: Use the NBIA Data Retriever software offered by TCIA for bulk downloads. It simplifies the process of managing and downloading large, complex collections, which can be cumbersome through the web interface alone.
| Feature | Details |
|---|---|
| Data Types | CT, MRI, PET, Digital Pathology, Genomics |
| Cost | Free (public access) |
| Annotations | Varies by collection; many include expert segmentations. |
| Best For | Oncology AI, Radiomics, Reproducible Research |
Website: https://www.cancerimagingarchive.net/
2. The Cancer Imaging Archive (TCIA)
The Cancer Imaging Archive (TCIA) is an indispensable, publicly funded resource for anyone working in oncology AI. It serves as a large-scale repository of de-identified medical images, primarily focused on cancer, and is an essential starting point for training and validating diagnostic algorithms. What sets TCIA apart is its rich, multi-modal data integration; images are often linked with corresponding clinical outcomes, genomic data, and pathology reports. This provides a holistic view crucial for developing sophisticated predictive models from a diverse medical image dataset.

Key Features and Use Cases
The platform is more than just a data dump; it’s a well-structured ecosystem for reproducible research. The data is standardized in DICOM format, ensuring interoperability and simplifying data processing pipelines for medical researchers and technology companies.
- Ideal Use Case: Excellent for training computer-aided detection (CADe) and diagnosis (CADx) systems for various cancers, such as lung, breast, and brain tumors. The associated clinical data supports projects aiming to predict patient prognosis or treatment response from imaging features.
- Access Requirements: While the majority of datasets are freely and publicly accessible, some collections are restricted and require an application to protect patient privacy or due to specific use agreements.
- Practical Tip: Use the NBIA Data Retriever software offered by TCIA for bulk downloads. It simplifies the process of managing and downloading large, complex collections, which can be cumbersome through the web interface alone.
| Feature | Details |
|---|---|
| Data Types | CT, MRI, PET, Digital Pathology, Genomics |
| Cost | Free (public access) |
| Annotations | Varies by collection; many include expert segmentations. |
| Best For | Oncology AI, Radiomics, Reproducible Research |
Website: https://www.cancerimagingarchive.net/
Btw, we build web DICOM viewers and custom medical imaging CRMs for our clients. Check out our portfolio.
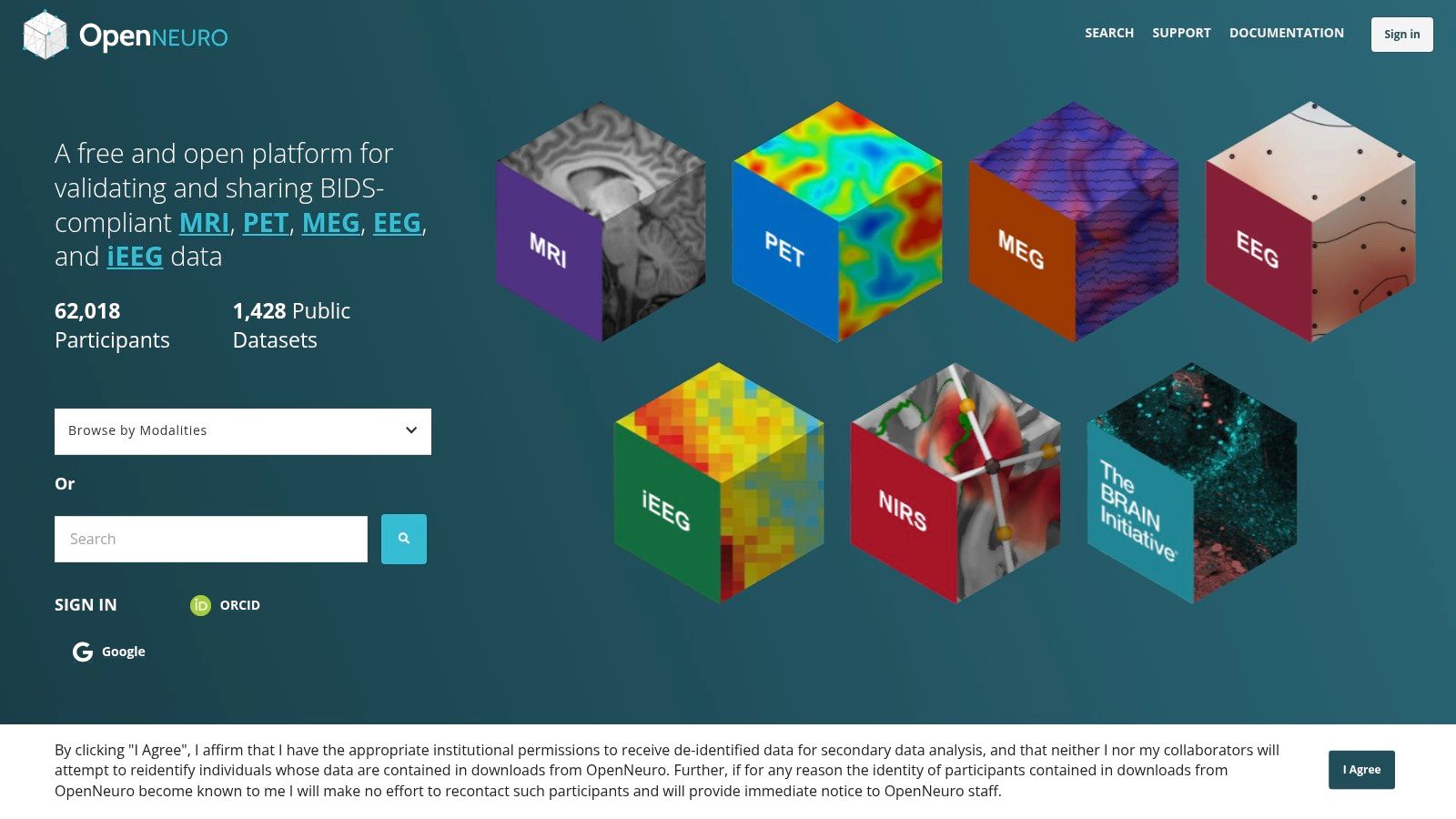
3. OpenNeuro
OpenNeuro is a cornerstone for the neuroscience community, functioning as an open-science platform dedicated to sharing human brain imaging data. Its primary mission is to foster reproducibility and transparency in research by hosting a vast collection of neuroimaging datasets. What truly distinguishes OpenNeuro is its strict adherence to the Brain Imaging Data Structure (BIDS) standard, a community-driven specification for organizing and describing neuroimaging data. This standardization simplifies data reuse and makes it an invaluable medical image dataset resource for large-scale meta-analyses and validation studies.
Key Features and Use Cases
The platform is designed to make neuroimaging data findable, accessible, interoperable, and reusable (FAIR). It features a user-friendly interface for browsing and downloading datasets directly in their standardized BIDS format.
- Ideal Use Case: Perfect for researchers studying brain function, structure, and connectivity. It’s an excellent source for training algorithms on tasks like brain segmentation, functional MRI (fMRI) analysis, and EEG signal processing.
- Access Requirements: All public datasets are completely free and open to access without any registration, although users can create an account to upload their own data.
- Practical Tip: Leverage the platform’s built-in filtering and search capabilities to quickly find datasets by modality (e.g., MRI, EEG), task (e.g., resting-state, memory), or subject count. This saves significant time compared to manually browsing the extensive collection.
| Feature | Details |
|---|---|
| Data Types | MRI, PET, MEG, EEG, iEEG |
| Cost | Free (public access) |
| Annotations | Varies; metadata is standardized according to BIDS. |
| Best For | Neuroscience Research, Brain Mapping, Reproducibility Studies |
Website: https://openneuro.org/
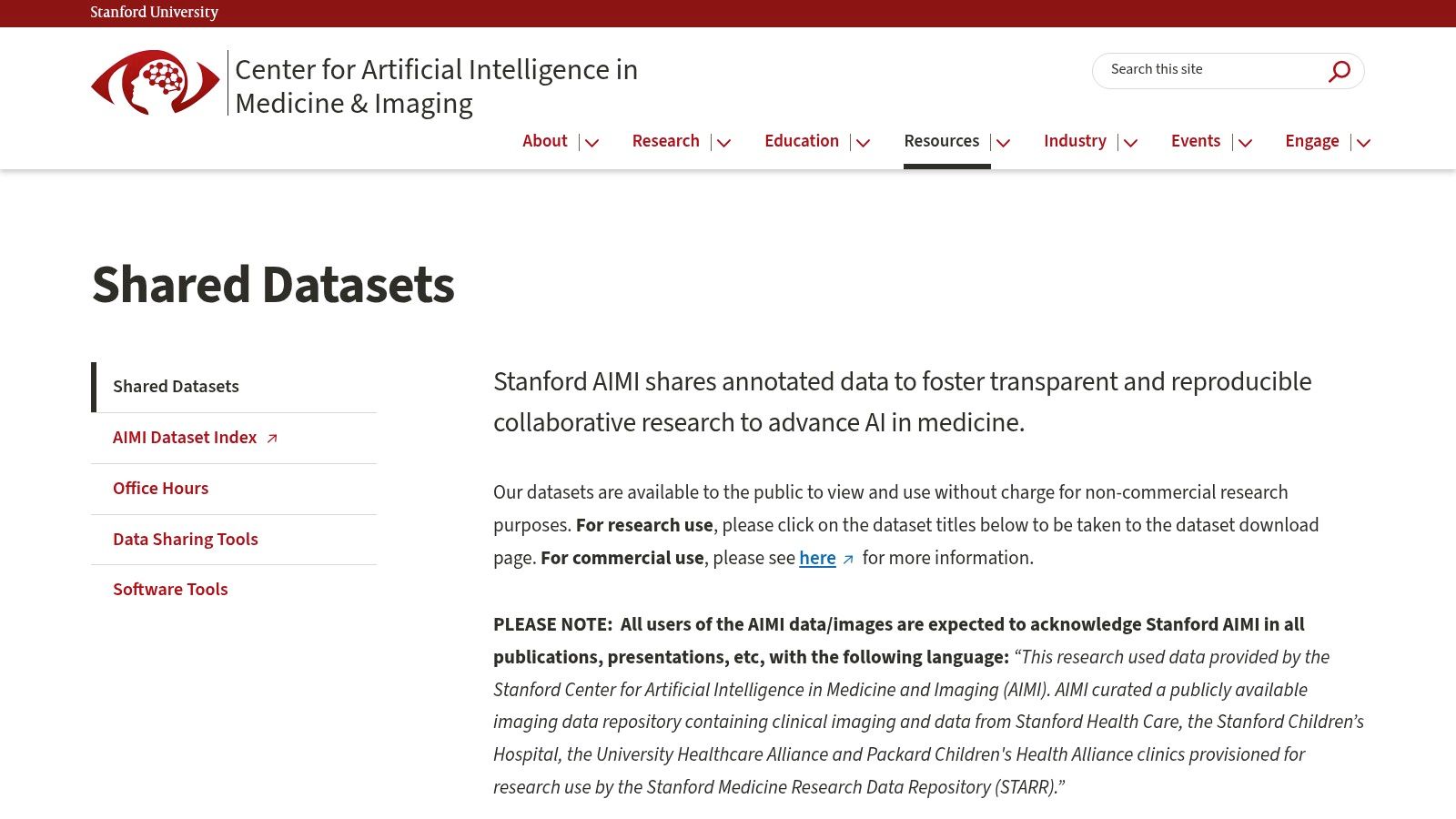
4. Stanford AIMI Center Datasets
The Stanford Center for Artificial Intelligence in Medicine & Imaging (AIMI) provides a curated collection of high-quality, expert-annotated clinical imaging datasets. Sourced primarily from Stanford Health Care, these datasets are specifically designed to accelerate AI research and development. What distinguishes the AIMI repository is its focus on providing clean, well-documented data across diverse modalities, including radiographs, CT scans, and echocardiograms, lowering the barrier to entry for researchers looking to validate their models on real-world clinical data.
Key Features and Use Cases
The center emphasizes transparent and reproducible research by providing detailed documentation and, in many cases, the original publications associated with each dataset. This makes it an invaluable educational and benchmarking tool.
- Ideal Use Case: Perfect for projects requiring meticulously annotated data for tasks like disease classification in chest X-rays (CheXpert), abnormality detection in musculoskeletal radiographs (MURA), or segmentation in brain CT scans (CQ500).
- Access Requirements: Access is free for non-commercial research purposes. Users must agree to a data use agreement for each dataset, which outlines usage restrictions and attribution requirements. Commercial use requires a separate license.
- Practical Tip: Pay close attention to the “Known Issues” or “Limitations” sections provided for each dataset. This transparency helps researchers anticipate potential biases or challenges and design more robust experiments.
| Feature | Details |
|---|---|
| Data Types | Radiographs (X-Ray), CT, Echocardiograms, MRI |
| Cost | Free (non-commercial research) |
| Annotations | High-quality expert labels and segmentations. |
| Best For | Benchmarking Models, Educational Use, Clinical AI Validation |
Website: https://aimi.stanford.edu/shared-datasets
5. UK Biobank
UK Biobank is a monumental, large-scale biomedical database and research resource, containing in-depth genetic and health information from half a million UK participants. Its unique strength lies in linking this extensive clinical and genomic data with a massive and growing repository of medical images, including brain, cardiac, and abdominal MRIs. This makes it an unparalleled resource for studying the complex interplay between genetics, lifestyle, and disease manifestation visible through imaging.
Key Features and Use Cases
The power of UK Biobank is its sheer scale and multi-modal integration, enabling population-level studies that are otherwise impossible. Researchers can explore how early imaging markers correlate with future health outcomes across a vast cohort.
- Ideal Use Case: Perfect for large-scale epidemiological studies, identifying novel imaging biomarkers for neurodegenerative diseases like dementia, or understanding cardiovascular risk factors. It’s a goldmine for any medical image dataset project linking imaging phenotypes to genetic predispositions.
- Access Requirements: Access is not open; it requires a formal application process to be reviewed and approved. Researchers must demonstrate a valid health-related research interest. There are also access fees associated with using the data.
- Practical Tip: The application process is rigorous. Before applying, thoroughly explore the UK Biobank Data Showcase to understand the exact variables and imaging data available to ensure it aligns with your research question.
| Feature | Details |
|---|---|
| Data Types | MRI (brain, cardiac, abdominal), DEXA scans, Genomics, Health Records |
| Cost | Application and access fees apply |
| Annotations | Basic segmentations and derived imaging phenotypes are often available. |
| Best For | Population Imaging, Epidemiological Studies, GWA Studies |
Website: https://www.ukbiobank.ac.uk/
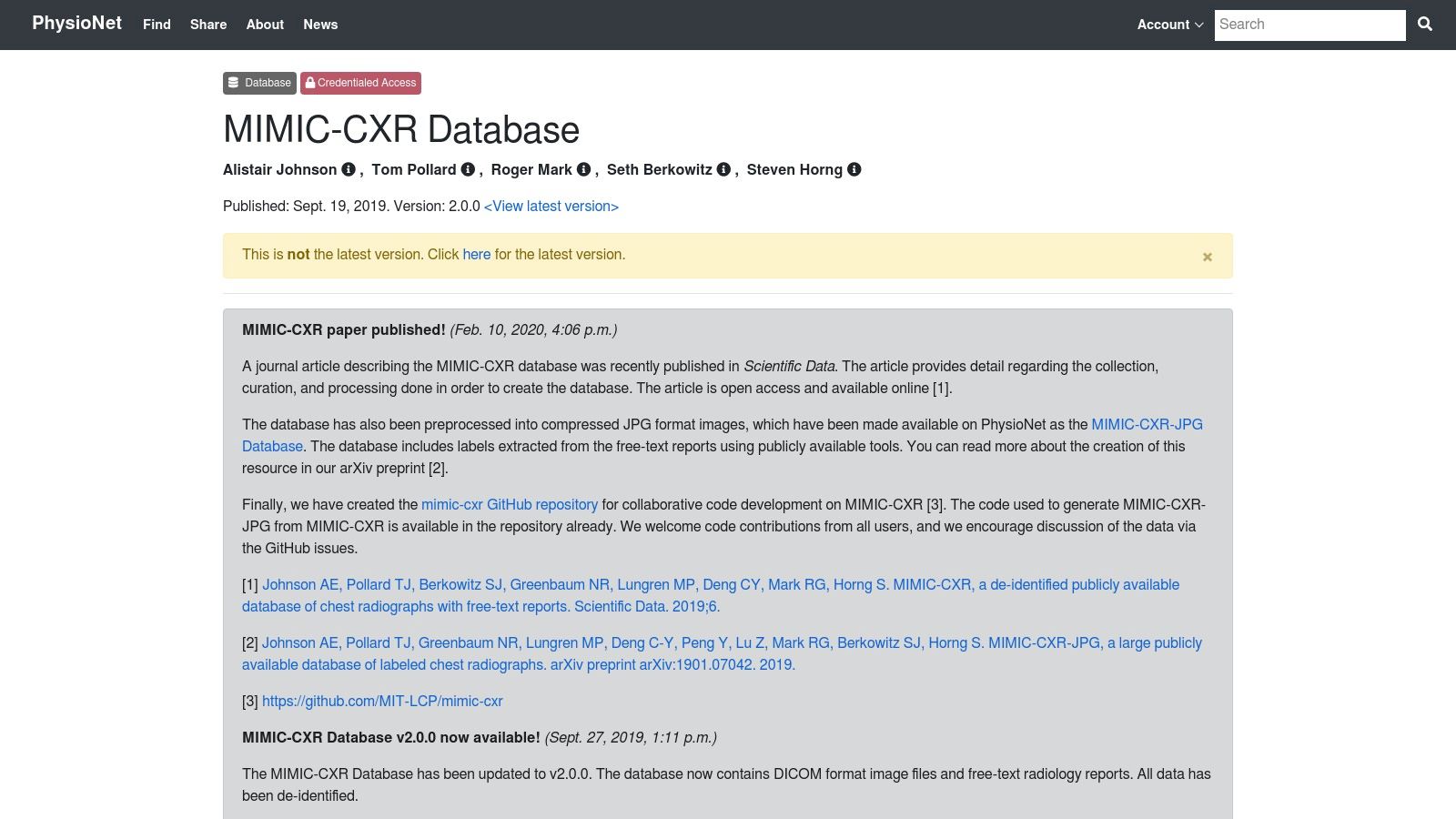
6. MIMIC-CXR Database
The MIMIC-CXR Database is a cornerstone resource for developing AI in thoracic imaging. Hosted on PhysioNet, this large-scale, publicly available medical image dataset contains over 377,000 de-identified chest radiographs. What truly distinguishes MIMIC-CXR is its powerful multi-modal nature; each DICOM image is directly linked to a corresponding free-text radiology report. This unique combination allows researchers to bridge the gap between pixel data and clinical interpretation, enabling the development of advanced models that can both identify findings and generate descriptive reports.
Key Features and Use Cases
The database is meticulously curated for research, with comprehensive metadata accompanying the images, all within a de-identified and publicly accessible framework. It’s an ideal playground for natural language processing (NLP) and computer vision tasks.
- Ideal Use Case: Perfect for training models that perform automated chest X-ray interpretation and report generation. It’s also excellent for developing systems that can classify pathologies based on both the image and the associated radiologist’s notes.
- Access Requirements: Access is free but requires completing a credentialing process on PhysioNet, which involves a short training course on human subjects research to ensure data is used responsibly.
- Practical Tip: Leverage the structured labels file (
mimic-cxr-2.0.0-chexpert.csv.gz) provided with the dataset. This file contains 14 common chest radiographic observations extracted from the reports, which can be used as ground-truth labels to jumpstart classification model training without needing to process the raw text yourself.
| Feature | Details |
|---|---|
| Data Types | Chest X-ray (Radiographs), Free-text Radiology Reports |
| Cost | Free (requires credentialing) |
| Annotations | Structured labels for 14 common observations derived from reports. |
| Best For | Automated Radiology Reporting, Multi-modal Learning, NLP in Medicine |
Website: https://physionet.org/content/mimic-cxr/2.0.0/
7. MedPix Database
The MedPix Database is a powerful, open-access resource managed by the U.S. National Library of Medicine (NLM), designed for both educational and research applications. It stands out due to its case-based approach, presenting over 59,000 images linked to more than 12,000 patient cases. Each case is a mini-lesson, often including patient history, imaging findings, and diagnoses. This narrative context makes it an exceptional tool for training AI models to recognize not just image patterns, but also their clinical relevance, offering a unique type of medical image dataset.
Key Features and Use Cases
MedPix excels as a teaching file and a source for building versatile diagnostic algorithms. Its strength lies in the breadth of its topics and the detailed, searchable metadata that accompanies each image.
- Ideal Use Case: Excellent for developing and testing differential diagnosis algorithms. It’s also highly suitable for creating educational content or training junior radiologists and medical students on case interpretation across numerous specialties.
- Access Requirements: The entire database is completely free and open to the public. No registration is required, which significantly lowers the barrier to entry for researchers and educators.
- Practical Tip: Leverage the “Topic Search” feature to find cases related to specific diseases or anatomical regions. For more complex queries, the advanced search allows you to filter by patient age, gender, imaging modality, and findings.
| Feature | Details |
|---|---|
| Data Types | CT, MRI, X-ray, Ultrasound, Angiography, Nuclear Medicine |
| Cost | Free (public access) |
| Annotations | Annotations are provided as case descriptions and findings, not pixel-level masks. |
| Best For | Medical Education, Case-Based Learning, Differential Diagnosis AI |
Website: https://medpix.nlm.nih.gov/
8. NIH Chest X-Ray Dataset
The NIH Chest X-Ray Dataset is a landmark public resource that significantly advanced the field of deep learning in medical diagnostics. It contains over 112,000 frontal-view chest X-ray images from more than 30,000 unique patients, making it one of the largest and most widely cited public chest X-ray collections. Its major contribution lies in its disease labels, which were extracted from associated radiological reports using natural language processing (NLP). This approach provided a massive, albeit imperfect, labeled medical image dataset for developing disease detection algorithms.
Key Features and Use Cases
The dataset’s scale makes it a go-to for benchmarking and pre-training models for thoracic pathology classification. The images are provided in PNG format, making them easily accessible for researchers without specialized DICOM software.
- Ideal Use Case: Excellent for training and validating automated systems to detect common thoracic diseases like pneumonia, pneumothorax, and nodules. It’s a foundational dataset for projects focused on multi-label classification from chest radiographs.
- Access Requirements: The dataset is completely free and open for public access. No registration or application is needed, allowing for immediate download and use.
- Practical Tip: The NLP-derived labels can be noisy. Researchers often use techniques like label smoothing or develop consensus from multiple models to mitigate the impact of potential inaccuracies in the original report-based labels.
| Feature | Details |
|---|---|
| Data Types | Chest X-ray (PNG) |
| Cost | Free (public access) |
| Annotations | 14 common thoracic disease labels derived via NLP. |
| Best For | Chest pathology classification, Benchmarking models, Pre-training |
Website: https://nihcc.app.box.com/v/ChestXray-NIHCC
9. MURA (Musculoskeletal Radiographs) Dataset
The MURA (Musculoskeletal Radiographs) dataset, developed by the Stanford ML Group, is one of the largest public radiographic image collections available. It provides a massive trove of over 40,000 musculoskeletal X-rays of the upper extremities, including studies of the shoulder, humerus, elbow, forearm, wrist, hand, and finger. Each study was manually labeled by board-certified radiologists as either normal or abnormal, making it an invaluable resource for binary classification tasks. MURA’s scale and focus make it a benchmark medical image dataset for developing and testing automated diagnostic systems in orthopedics.
Key Features and Use Cases
The dataset was the basis of a competition to see if AI models could outperform radiologists at detecting abnormalities, a testament to its quality and challenging nature. The data is organized by body part, providing a clean structure for targeted model training.
- Ideal Use Case: Perfect for building and validating deep learning models for abnormality detection in musculoskeletal X-rays. It’s also suitable for research into transfer learning, model generalization across different anatomical regions, and explainable AI (XAI) in radiology.
- Access Requirements: Access is free but requires signing a dataset usage agreement to ensure the data is used for research purposes only. Once approved, the dataset can be downloaded directly.
- Practical Tip: The normal/abnormal labels are at the study level, not the image level. A study can contain multiple images, so be sure to aggregate predictions correctly when evaluating your model’s performance against the provided ground truth.
| Feature | Details |
|---|---|
| Data Types | Digital Radiographs (X-ray) |
| Cost | Free (requires user agreement) |
| Annotations | Study-level labels (normal vs. abnormal) by radiologists. |
| Best For | Orthopedic AI, Binary Classification, Anomaly Detection |
Website: https://stanfordmlgroup.github.io/competitions/mura/
10. re3data (Registry of Research Data Repositories)
Unlike platforms that host data directly, re3data serves as a comprehensive global registry of research data repositories. It’s an invaluable discovery tool, a “meta-repository” that helps researchers locate the perfect medical image dataset from thousands of sources worldwide. Instead of hosting images, it provides detailed, structured information about other repositories, including their subject matter, access policies, and data standards. This makes it an essential first stop for broadening your search beyond the most well-known archives.
Key Features and Use Cases
The power of re3data lies in its extensive search and filtering capabilities, allowing users to efficiently navigate the vast landscape of data repositories to find what they need.
- Ideal Use Case: Excellent for exploratory research when you need to find a niche or highly specific medical image dataset that might not be available on larger, more generalized platforms. It’s also perfect for verifying the credibility and policies of a repository you’ve discovered elsewhere.
- Access Requirements: Varies entirely by the listed repository. re3data clearly indicates the access type (open, restricted, closed) for each entry, but the user must follow the specific requirements of the ultimate data source.
- Practical Tip: Use the advanced search filters to narrow down repositories by “Content Types” (e.g., “Images”) and “Subjects” (e.g., “Medicine,” “Neurosciences”). This quickly isolates relevant sources from the thousands of entries.
| Feature | Details |
|---|---|
| Data Types | A registry covering all data types, including CT, MRI, X-Ray, and more. |
| Cost | Free to use the registry. |
| Annotations | Varies by the individual repository listed. |
| Best For | Discovering new and niche datasets, Verifying repository credentials. |
Website: https://www.re3data.org/
11. Nightingale Open Science
Nightingale Open Science is a collaborative platform dedicated to advancing AI in medicine by providing access to high-quality, ground-truth-labeled medical imaging data. It partners with global health systems to curate and de-identify extensive datasets, making them available on secure cloud infrastructure. The platform’s core mission is to empower non-profit research, removing the significant barrier of data acquisition and allowing researchers to focus on developing and validating new algorithms for a wide range of medical conditions.
Key Features and Use Cases
The strength of Nightingale lies in its commitment to providing well-curated, labeled data, which is often a major bottleneck in AI development. This focus on quality and accessibility makes it a valuable resource for the academic and non-profit sectors.
- Ideal Use Case: Perfect for academic labs or non-profit organizations developing AI models for diagnostics, particularly when ground-truth labels are essential for supervised learning. It’s well-suited for projects targeting conditions beyond oncology.
- Access Requirements: Access is restricted to non-profit research. Users must complete a registration and approval process to gain access to any medical image dataset, ensuring data is used ethically and for its intended purpose.
- Practical Tip: When applying for access, be very clear and specific about your research proposal and how the data will be used. A well-defined project plan increases the likelihood of a swift approval.
| Feature | Details |
|---|---|
| Data Types | Varies by collection; focuses on diverse conditions. |
| Cost | Free (for approved non-profit research) |
| Annotations | High-quality, ground-truth labels are a key feature. |
| Best For | Academic AI Research, Supervised Learning, Cross-Institutional Collaboration |
Website: https://www.nightingalescience.org/
12. Medical Open Network for AI (MONAI)
Medical Open Network for AI (MONAI) is less of a direct medical image dataset provider and more of a powerful open-source framework built to accelerate AI in healthcare. It’s a PyTorch-based toolkit that provides domain-optimized, standardized tools for every stage of the deep learning workflow. What makes MONAI a critical resource is its integrated access to various public datasets and pre-built pipelines, effectively lowering the barrier to entry for developing and validating medical imaging models. It bridges the gap between research and deployment with tools designed for reproducibility.
Key Features and Use Cases
MONAI provides a cohesive ecosystem of data loaders, transformations, and network architectures specifically for medical imaging. This specialization ensures that common challenges, like handling 3D data or diverse imaging formats, are addressed out of the box.
- Ideal Use Case: Perfect for researchers and developers who need a robust, reproducible environment for building, training, and evaluating models. It is especially useful for tasks like 3D segmentation, registration, and classification across various modalities.
- Access Requirements: The framework and its core tools are completely free and open-source. Access to specific datasets through MONAI depends on the original dataset’s license and access policies.
- Practical Tip: Leverage the MONAI Label tool. It’s an intelligent image labeling and learning tool that can significantly speed up the annotation process by using AI-assisted methods, turning a tedious task into a semi-automated one.
| Feature | Details |
|---|---|
| Data Types | Framework supports CT, MRI, Ultrasound, Pathology |
| Cost | Free (Open-source) |
| Annotations | Provides tools for creating annotations; not a source of pre-annotated data itself. |
| Best For | Reproducible AI Research, Model Development, Annotation |
Website: https://monai.io/
12 Medical Image Dataset Comparison
| Product / Dataset | Core Features / Modality | User Experience / Quality ★★★★☆ | Value Proposition 💰 | Target Audience 👥 | Unique Selling Points ✨ |
|---|---|---|---|---|---|
| Free AI Medical Imaging Annotation | CT scan; AI-powered organ segmentation (6 organs) | Easy web interface, accurate 3D models | Free access to advanced AI tools | Medical professionals & researchers | Instant 3D DICOM upload; zero installation |
| The Cancer Imaging Archive (TCIA) | Multi-modal (CT, MRI, PET); cancer focus | Standardized, wide-ranging datasets | Free, regularly updated | Cancer researchers & AI developers | Clinical/genomic data integration |
| OpenNeuro | Multi-modal neuroimaging (MRI, PET, EEG etc.) | Community-driven, reproducible data | Free, open access | Neuroscientists & AI researchers | Large neuro dataset; community standards |
| Stanford AIMI Center Datasets | Multi-modal clinical imaging; annotated | High-quality, well-annotated | Free for non-commercial research | AI researchers & clinicians | Institutional multi-source data |
| UK Biobank | MRI, CT + genetics & health records | Large cohort, comprehensive | Access with approval, potential fees | Epidemiologists & large-scale research | Multi-modal linked data |
| MIMIC-CXR Database | Chest X-ray + free-text radiology reports | Large-scale, de-identified | Free access | Radiology AI researchers | Multi-modal imaging + NLP reports |
| MedPix Database | Diverse specialties; 59k+ images | Searchable metadata, educational | Free, no registration | Medical educators & researchers | Organized by anatomy & pathology |
| NIH Chest X-Ray Dataset | Chest X-ray; labeled via NLP | Large, labeled dataset | Free access | AI model developers & researchers | Largest public labeled chest X-ray dataset |
| MURA (Musculoskeletal Radiographs) | Musculoskeletal radiographs; labeled normal/abnormal | High-quality, radiologist labeled | Free with registration | Musculoskeletal researchers & AI | Large labeled radiograph dataset |
| re3data (Data Repositories Registry) | Registry of medical & other repositories | Centralized directory, updated | Free directory, no data hosting | Researchers seeking datasets | Broad multi-discipline registry |
| Nightingale Open Science | Curated, labeled datasets on cloud | High-quality, ground-truth labeled | Free for non-profit research | Non-profit AI researchers | Collaborative global health system partnerships |
| Medical Open Network for AI (MONAI) | Open-source deep learning + dataset access | Community-supported, tool-rich | Free, continuous updates | AI developers and researchers | Framework + datasets for reproducible AI |
13. More and more datasets!
SAROS dataset
This is a goldmine of 900 CT scans with detailed notes about different body parts and organs. It’s perfect for anyone working on medical image analysis and it’s totally free to use.
Paper: https://lnkd.in/ekQnxpNb
Dataset: https://lnkd.in/eZAUxRwf
TotalSegmentator from the NCI Imaging Data Commons
They’ve added features and segmentations for over 126,000 CT scans from the NLST collection. This is huge for AI research in medicine!
Discussion: https://lnkd.in/e3-wGehc
Dataset: https://lnkd.in/ebaE-dbC
Healthy whole body CT scans from the cancer imaging archive
It contains 30 CT scans of full body.
Dataset: https://lnkd.in/egVyuuGH
Histopathology and spatial transcriptomics
Repo: https://lnkd.in/enWfgtBh
Dataset: https://lnkd.in/ev__8tbU
BRACS: BReAst Carcinoma Subtyping
Dataset download page (requires registration): https://lnkd.in/e4FZQ7TY
BCBM-RadioGenomics | MRI Dataset of Metastatic Breast Cancer to the Brain with Expert-reviewed Segmentations and Tumor-derived Radiomic Features
Dataset download page: https://lnkd.in/eaekwCrK
TOMPEI-CMMD Dataset | The TOMPEI-CMMD dataset adds segmentations (masses, calcifications, etc) and corrections to labeling errors in the original Chinese Mammography Database (CMMD) previously published on TCIA
Dataset download page: https://lnkd.in/ejKSwZe5
Stroke dataset From Teknofest AI Healthcare competition
Dataset download page: https://lnkd.in/ezCRDY3T
A bunch of datasets from Osirix
Cardiac and coronary study, CTA abdomen and lower extremities,…
Datasets download page: https://lnkd.in/eJcAy5Dv
HaN-Seg dataset which includes CT and MR image pairs with manual segmentations of 30 organs
It’s well-suited for segmentation and image-to-image translation tasks.
Dataset download page: https://lnkd.in/eYs8gsTQ
Automated Segmentation of Coronary Arteries
Dataset download page: https://lnkd.in/eY64_6qU
Other datasets shared in the comments but I couldn’t find their download links:
Accelerating Your AI Journey with the Right Data and Tools
Navigating the landscape of medical imaging data is the foundational step in any AI development lifecycle. As we’ve explored, the resources available are vast and varied, ranging from highly specialized collections like The Cancer Imaging Archive (TCIA) for oncology to broad, multi-modal repositories such as the UK Biobank. Each medical image dataset serves a unique purpose, whether it’s for training a model to detect specific pathologies in chest X-rays using the MIMIC-CXR database or for developing novel neurological algorithms with data from OpenNeuro.
The journey, however, rarely ends with data acquisition. The transition from raw pixels to a clinically relevant, deployable AI solution is a complex process filled with critical milestones. Selecting the right dataset is just the beginning; the real work lies in preparing, annotating, and validating that data to suit your specific project needs.
Key Takeaways and Next Steps
To move forward effectively, consider these essential points:
- Define Your Goal First: Your specific clinical question or application dictates your data needs. A project aimed at musculoskeletal analysis will find immense value in the MURA dataset, whereas a general-purpose chest pathology detector might start with the NIH Chest X-Ray collection. Clearly defining your use case will prevent wasted effort on unsuitable datasets.
- Acknowledge Data Limitations: No public dataset is perfect. Be prepared to encounter challenges such as class imbalance, inconsistent annotation quality, or missing metadata. Acknowledge these limitations early and plan for data cleaning, pre-processing, and potentially supplementary annotation.
- Leverage Development Frameworks: The raw data is only one part of the equation. Tools like MONAI provide a standardized, PyTorch-based framework that can significantly streamline the entire development pipeline, from data loading and augmentation to training and validation. Integrating such frameworks can save your team hundreds of hours.
Btw, we build web DICOM viewers and custom medical imaging CRMs for our clients. Check out our portfolio.






