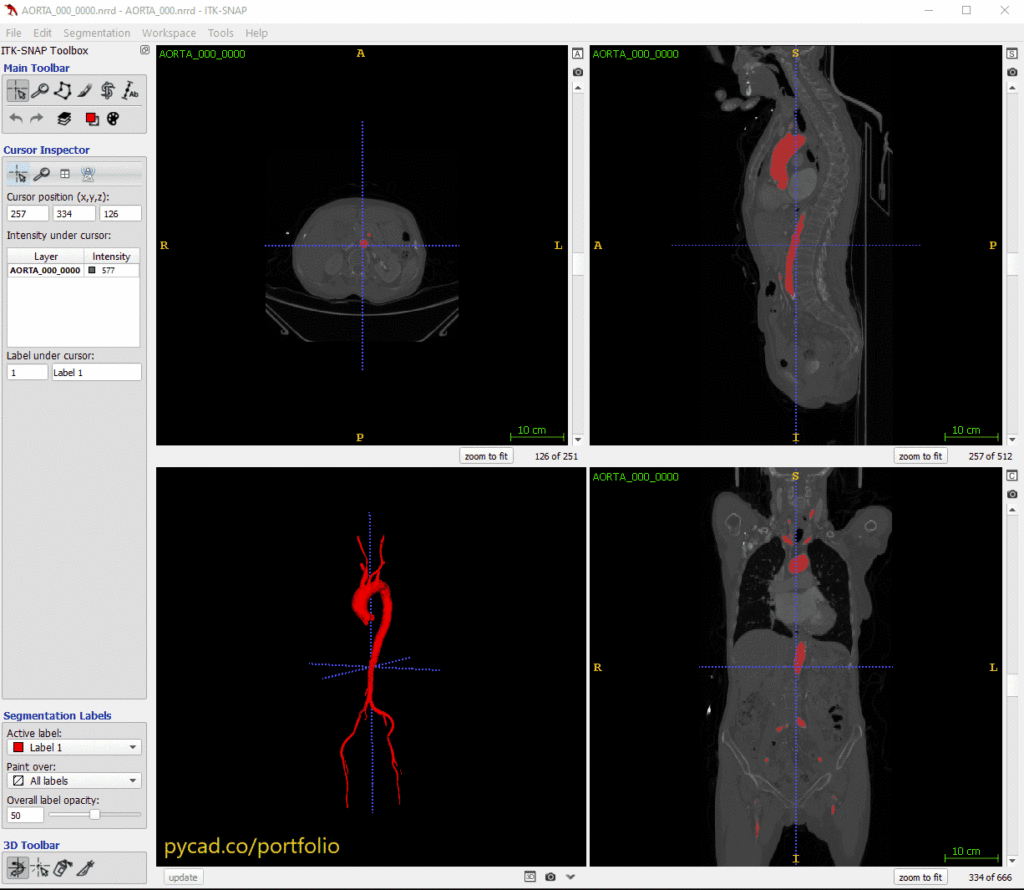
A complete walkthrough on how to use nnUNet to build powerful medical imaging segmentation models
Welcome to our deep dive into nnUNetv2, the renowned neural network framework specifically designed for medical image segmentation. Whether you’re a seasoned researcher or just starting your journey in medical imaging, this guide will try to give you the knowledge and tools to harness the full potential of nnUNet.
Note: I will be using the terms nnUNet and nnUNetv2 interchangeably. But in reality, I am covering the latest version of nnUNet, namely version 2.
Hardware requirements
In order to be able to use nnUNet effectively, you’ll need to know the following:
In the official documentation, they mention that they support GPU (recommended), CPU and Apple M1/M2 as devices.
However, here they are only mentioning the overall devices where you can run the code, but training a nnUNet model on a CPU is not really feasible. It would take ages. Even on a small dataset.
They talk about specific hardware requirements for training and inference on the same page. I will paste it here to make it easier for you! But if you want more details then I recommend you go back to the source!
Hardware Requirements for Training
“We recommend you use a GPU for training as this will take a really long time on CPU or MPS (Apple M1/M2). For training a GPU with at least 10 GB (popular non-datacenter options are the RTX 2080ti, RTX 3080/3090 or RTX 4080/4090) is required. We also recommend a strong CPU to go along with the GPU. 6 cores (12 threads) are the bare minimum! CPU requirements are mostly related to data augmentation and scale with the number of input channels and target structures. Plus, the faster the GPU, the better the CPU should be!”
Hardware Requirements for Inference
“Again we recommend a GPU to make predictions as this will be substantially faster than the other options. However, inference times are typically still manageable on CPU and MPS (Apple M1/M2). If using a GPU, it should have at least 4 GB of available (unused) VRAM.”
Example hardware configurations
Example workstation configurations for training:
CPU: Ryzen 5800X — 5900X or 7900X would be even better! We have not yet tested Intel Alder/Raptor lake but they will likely work as well.
GPU: RTX 3090 or RTX 4090
RAM: 64GB
Storage: SSD (M.2 PCIe Gen 3 or better!)
Example Server configuration for training:
CPU: 2x AMD EPYC7763 for a total of 128C/256T. 16C/GPU are highly recommended for fast GPUs such as the A100!
GPU: 8xA100 PCIe (price/performance superior to SXM variant + they use less power)
RAM: 1 TB
Storage: local SSD storage (PCIe Gen 3 or better) or ultra fast network storage
Setting up your Python environment
Before you can use nnUNet, you need to first set up a few things on your local machine or on a capable server.
Creating a virtual environment
Although not necessary, I highly recommend you create a virtual environment and install the necessary nnUNet packages inside. I would not recommend you install these packages directly on your system. This can ensure that in case things go wrong, you can simply delete the virtual environment and start again.
I personally prefer Miniconda but you can choose any other virtual environments handler that you like, such as: venv, virtualenv, or even Docker.
Installing Pytorch
Once your virtual environment is set up and activated, you should install Pytorch. In fact, nnUNet is fully based on Pytorch so it’s a key component to have.
Then you need to install nnUNet V2 package. You can achieve this in 2 different ways:
- For use as a standardized baseline: this means that you mainly want to be able to run commands from nnUNetv2 for training, evaluation and inference. If you’re trying to test nnUNetv2 for the first time, then this approach is for you. You can install nnUNetv2 as a standardized baseline by doing:
pip install nnunetv2
For use as an integrative framework: this means that you want to use nnUNet as part of your code. This is especially useful when you’re trying to build your custom inference pipeline, which uses nnUNet. If you choose this second option, then you can install it like this:
git clone https://github.com/MIC-DKFZ/nnUNet.git cd nnUNet pip install -e .
Then, you need to set paths on your machine which would represent where you save raw data, preprocessed data and trained models. To do this, you need to set 3 environment variables
- nnUNet_raw: this is where you put your starting point data. It should be data that follows the nnUNet datasets naming convention. We will explain this in more details in the next section.
- nnUNet_preprocessed: this is a folder where the preprocessed data will be saved. This folder will be used by nnUNet when you run the preprocessing command (we will cover it below) in order to save the preprocessed data.
- nnUNet_results: this is the folder where nnUNet will save the training artifacts, including model weights, configuration JSON files and debug output.
You can set these environment variables in a number of ways. But the easiest option would be to set them directly in the terminal where you’re going to run nnUNet commands. For example, for the environment variable nnUNet_raw on a Linux machine, you would do something like this:
export nnUNet_raw="/path/to/your/nnUNet_raw" # on Linux set nnUNet_raw=C:\path\to\your\nnUNet_raw # on Windows
Btw, this only sets the environment variables temporarily. Meaning that if you close your terminal or you open another terminal window, then these variables will need to be set again.
If you want a more permanent solution, then you should add these commands in the .bashrc file if you’re on Linux or you could add them as environment variables in the “Edit system environment variables” panel on Windows.
Preparing Your Dataset with nnUNet
First things first, your journey with nnUNet begins with dataset preparation. The framework expects datasets in a very specific format, inspired by the Medical Segmentation Decathlon. This structure is crucial in order to be able to use nnUNet.
Btw, if you are working with the Medical Segmentation Decathlon (MSD) dataset, transitioning to the nnUNet format is straightforward, thanks to the nnUNetv2_convert_MSD_dataset command. For more details about how to use the command, I recommend you run it with the help argument like this:
nnUNetv2_convert_MSD_dataset -h
This will show you all the necessary arguments that you need to pass to this command in order to transform your MSD dataset into nnUNet dataset format.
On the other hand, if you you’re working with any dataset other than MSD, then you need to organize and name your dataset in the nnUNet format. Here’s how.
Before we go any further, your dataset should have the images and the segmentation. The images would be the medical scans (CT, MRI, ..) and the segmentations should be the annotated files made by expert medical professionals.
In the next paragraphs, I will describe the different folders and files you need in order to train a nnUNet model. But first, you should know that nnUNet expects the main folder, which would contain all of the training and testing data, to follow a specific format. The format is Dataset[IDENTIFIER]_[Name]. The IDENTIFIER is a number of your choosing. It should be different than other numbers that nnUNet has already assigned to other datasets and it should have 3 digits. The Name is just a name that you can give to your dataset. For example, for a dataset that I personally used to segment the aorta, my folder name was Dataset300_Aorta.
Now, inside this main folder, you should have a folder of training images called imagesTr and a folder containing your training labels (segmentations) called labelsTr. Optionally, you can add a test folder containing testing images, which should be called imagesTs. You don’t need to have labels for your testing images.
Your images should have the format:
{CASE_IDENTIFIER}_{XXXX}.{FILE_ENDING}
The CASE_IDENTIFIER is just a unique name for the dataset, which should contain a unique identifier number. For example, one of my training images (CT scan) is named AORTA_000_0000.nrrd.
And the segmentations should have the following format:
{CASE_IDENTIFER}.{FILE_ENDING}
An example of this from my aorta dataset is AORTA_000.nrrd.
Finally, you should have a json file called dataset.json. This file should contain meta-information about your dataset. Here’s an example from my aorta dataset:
{
"channel_names": {
"0": "CT"
},
"labels": {
"background": 0,
"AORTA": 1
},
"numTraining": 51,
"file_ending": ".nrrd",
"overwrite_image_reader_writer": "SimpleITKIO"
}
As you can see, in this file, I am specifying channel_names which is just CT in our case. I am also specifying labels, which are 2 in my case, the background and the aorta. There is also numTraining, which is the number of your training images (scans) and there is file_ending which is the extension. In my case it’s .nrrd.
The final key, overwrite_image_reader_writer, is optional. In my case, since I will be reading .nrrd files, I kept it as SimpleITKIO. You can omit this key and nnUNet will figure out the best I/O tool to read and write your data.
By now, you should have your main folder container several subfolder as well as this meta information json file. Here’s a screenshot of my own folder:
The main folder, Dataset300_Aorta, needs to be put inside the nnUNet_raw folder path. Now, you’re ready to go to the next steps, which are preprocessing and then training.
Start the Training
Training nnUNet models is a multi-step process, beginning with preprocessing your dataset. This crucial step, is accomplished with the nnUNetv2_plan_and_preprocess command. This command will preprocess the data that was put in nnUNet_raw folder path and it will save the preprocessed data to the nnUNet_preprocessed folder path. Here’s an example of how to run this command:
nnUNetv2_plan_and_preprocess -d DATASET_ID --verify_dataset_integrity -np 1
DATASET_ID here would be the number or identifier you chose for your dataset. In my case, it’s 300.
-np: the number of processes or workers to run your preprocessing. Putting a high value can kill the whole process (it needs a big RAM!). In most of my cases, I had to set it to 1 to avoid the process being killed.
The actual training command, nnUNetv2_train, kicks off your model’s learning journey. With options to customize the training duration and model configuration, nnUNet provides a lot of flexibility. Here’s an example of how to run the training:
nnUNetv2_train 300 3d_fullres all -tr nnUNetTrainer_250epochs
300 is the dataset ID.
3d_fullres is the nnUNet config that we chose. There are 3 others: 2d, 3d_lowres, 3d_cascade_fullres.
all refers to the fact that we want to use the whole training folder for training instead of doing cross-validation.
-tr nnUNetTrainer_250epochs refers to a specific trainer config where we run the training for 250 epochs. This config can be chosen from a variety of configs found here. You can also create your own config and add it to that file.
Inference & Evaluation
Once training is complete, inference is your next step. Using the nnUNetv2_predict command, you can easily apply your trained model to new datasets and generate predictions. Here’s an example on how to do so:
nnUNetv2_predict -i nnUNet_dirs/nnUNet_raw/Dataset300_Aorta/imagesTs -o nnUNet_dirs/nnUNet_raw/nnUNet_tests/ -d 300 -c 3d_fullres -tr nnUNetTrainer_250epochs -f all
Evaluation is the final piece of the puzzle. With nnUNetv2_evaluate_folder, you can assess the performance of your model against ground truth annotations, giving you valuable insights into its accuracy and efficiency. Here’s an example of how to run the evaluation:
nnUNetv2_evaluate_folder /nnUNet_tests/gt/ /nnUNet_tests/predictions/ -djfile Dataset300_Aorta/nnUNetTrainer_250epochs__nnUNetPlans__3d_fullres/dataset.json -pfile Dataset300_Aorta/nnUNetTrainer_250epochs__nnUNetPlans__3d_fullres/plans.json
The first argument you give to this command is the ground truth folder.
The second argument is the predictions folder.
-djfile is the path to your dataset.json file.
-pfile is the path to the plans.json file, which was created during the preprocessing step.
When you run the evaluation command, you’ll get something like this:
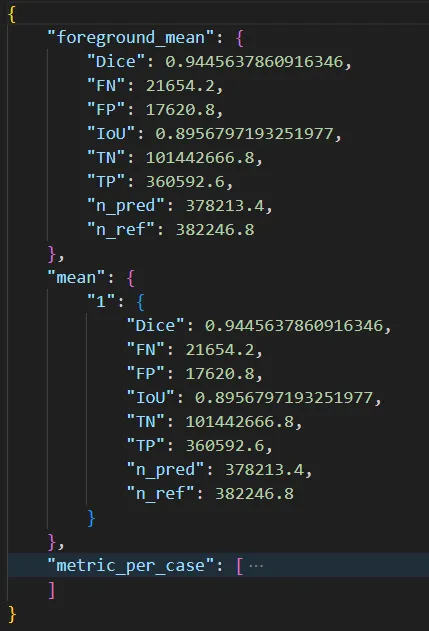
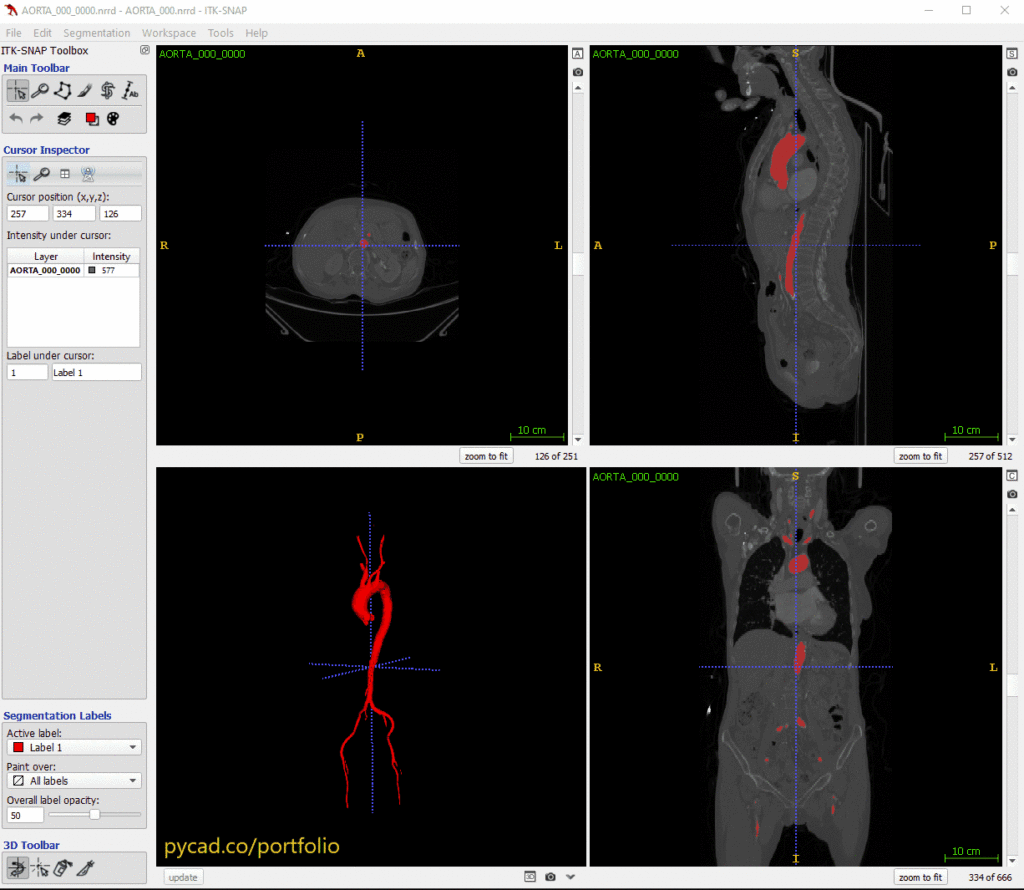
Here’s a sample output from our training of nnUNet on the Aorta dataset:
Conclusion
You have reached the end of this guide, congrats!
There is a ton of information that I could have added further to this guide. But I tried to keep it simple and straightforward and omit the details that could be cumbersome if you’re using nnUNet for the first time. For example, every command that I used in the guide, has a ton of extra arguments that allow you to tailor your training to your specific needs and on your specific hardware. The best way to explore these is to:
- Check the original documentation.
- Use the — help argument with any nnUNet command you’re using. It will give you thorough details about the different possibilities of that command.
- Check out also the issues opened in the original nnUNet GitHub repo. I found a lot of useful information there as well!
Btw, we at PYCAD are constantly building, training and deploying machine learning models for medical imaging applications. We have dealt with CT, MRI, CBCT and more! We have built models for segmenting different anatomical structures of the body (aorta, liver, lungs, hepatic blood vessels, tumors, mandible, maxilla, teeth, TMJs, sinuses, and more!). We have deployed these ML models locally and on the cloud through custom-made REST APIs or platform-specific endpoints like AWS SageMaker endpoints (synchronous or asynchronous). We can help you do the same!
If your startup or company is looking to work with people who have worked on several projects in the medical imaging field, then feel free to reach out to us at: contact@pycad.co. We do not work on student or research projects though.