Introduction
Segmentation in medical imaging is an important task that can help doctors diagnose and treat a wide range of conditions. Traditional segmentation methods can be time-consuming and require a lot of manual input. However, recent advances in machine learning have made it possible to automate this process and achieve impressive results. In this blog post, we’ll explore how the Segment Anything Model (SAM) can be adapted for medical imaging segmentation using DICOM files.
The Segment Anything Model (SAM)
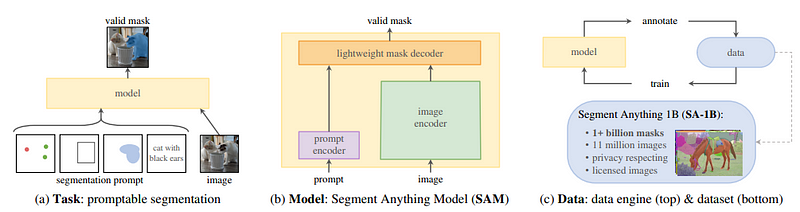
The Segment Anything Model (SAM) is a state-of-the-art image segmentation model that was introduced by Meta. The SAM model is designed to be promptable, which means that it can generalize to new image distributions and tasks beyond those seen during training. This capability is achieved through the use of prompt engineering, where hand-crafted text is used to prompt the model to generate a valid response for the task at hand. SAM has three main components: an image encoder, a flexible prompt encoder, and a fast mask decoder. The image encoder uses a pre-trained Vision Transformer (ViT) that is adapted to process high-resolution inputs. The prompt encoder can take in two sets of prompts: sparse prompts such as points, boxes, and text, and dense prompts such as masks. Finally, the mask decoder efficiently maps the image embedding, prompt embeddings, and an output token to a mask. With its promptable design, the SAM model can transfer zero-shot to new image distributions and tasks, making it a versatile tool for image segmentation tasks.
SAM for Dicoms
We adopted the model architecture proposed by Meta and forked their repository for our use. However, we had to make modifications to enable the model to handle DICOM files as the original repository only supports PNG or JPEG images. To achieve this, we created a custom function that reads DICOM files and pre-processes them into the appropriate format that can be used by the SAM model. Here is a code snippet showcasing our implementation.
def prepare_dicoms(dcm_file, show=False):
dicom_file_data = pydicom.dcmread(dcm_file).pixel_array
HOUNSFIELD_MAX = np.max(dicom_file_data)
HOUNSFIELD_MIN = np.min(dicom_file_data)
HOUNSFIELD_RANGE = HOUNSFIELD_MAX - HOUNSFIELD_MIN
dicom_file_data[dicom_file_data < HOUNDSFILD_MIN] = HOUNSFIELD_MIN
dicom_file_data[dicom_file_data > HOUNSFIELD_MAX] = HOUNSFIELD_MAX
normalized_image = (dicom_file_data - HOUNS_MIN) / HOUNSFIELD_RANGE
uint8_image = np.uint8(normalized*255)
opencv_image = cv2.cvtColor(uint8_image, cv2.COLOR_GRAY2BGR)
if show:
cv2_imshow(opencv_image) # for Google Colab
return opencv_image
Output
I ran the model on a lot of examples, including some challenging pathologies, and it performed very well. Here is an example:
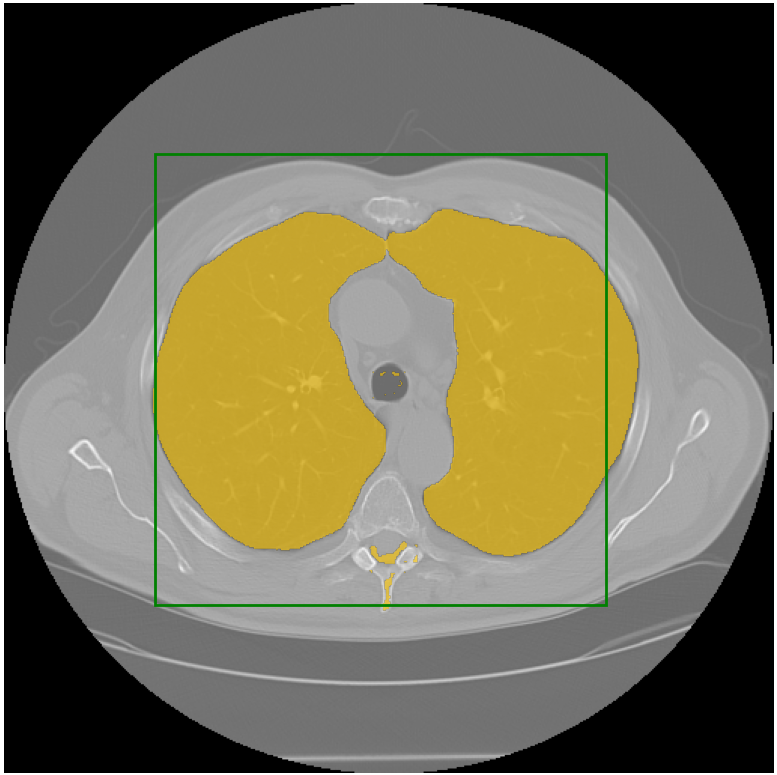
You can access the code for SAM applied to medical imaging on my repository at this link: https://github.com/amine0110/SAM-Medical-Imaging. Feel free to explore and test it for yourself.






