In this blog post, we will discuss the best libraries that can be used in Python for medical imaging.
Outline
- Pydicom
- Nibabel
- Dicom2nifti
- SimpleITK
- VTK
- Numpy to STL (using NumPy-stl)
Pydicom
Pydicom is an open-source library for working with Dicom files. It’s what we use to load, edit, and save Dicom files!
Examples:
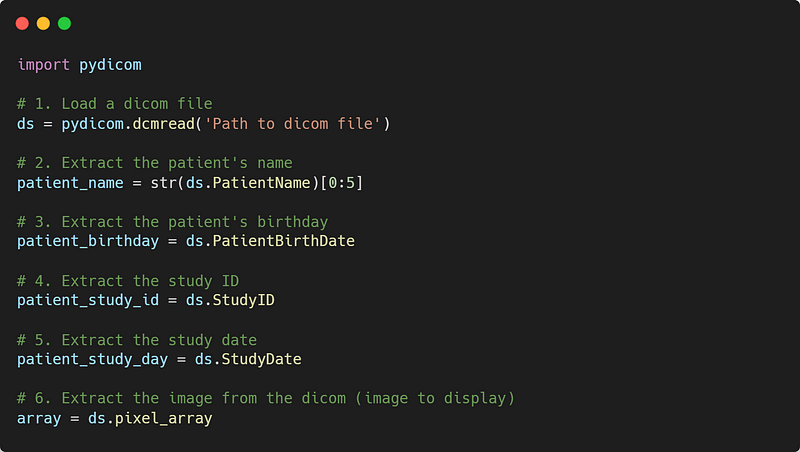
If we want to show/visualize the array extracted from the Dicom file, we can use the following code:

Nibabel
Then there’s nibabel, which is the most commonly used library for dealing with nifti files. There are many options in this library; here are a few to get you started:

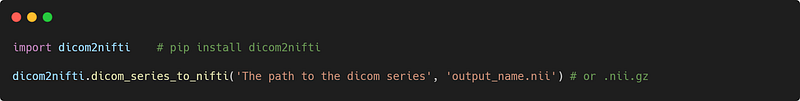
Dicom2nifti
This is a very useful library for converting Dicom series into nifti files with a single function.
SimpleITK
For me, simpleITK is the best library for treating or processing Dicom and nifti files because it contains all of the functions required for both formats and even offers options not found in other medical imaging libraries, such as converting nifti files into Dicoms series (this is one of the most sensitive conversions, and not available in other libraries).
Upload a nifti file
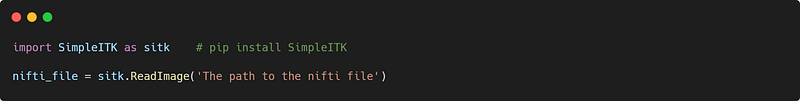
Lots of other options are available, you can check them out here.
VTK
Kitware provides VTK as a library. It includes not only conversion functions but also additional tools for visualization and medical imaging processing! This library is even used by some well-known software, such as 3D Slicer. This means that all of the features available in 3D Slicer can be found and used in VTK, which is fantastic.
Because there are so many things we can do with this library, I’ll provide a link to the documentation so you can directly check and find the exact functions/operations required for your projects!
Numpy-stl
This library is not only used for medical imaging, but it plays an important role in medical imaging by assisting in the creation of 3D meshes from segmentation (manual or automatic) or assisting in the conversion of nifti files into STLs, as discussed in this blog post.

MedPy
Another SimpleITK-based library for medical imaging that can be used for reading and writing files such as:
Medical formats:
- ITK MetaImage (.mha/.raw, .mhd)
- Neuroimaging Informatics Technology Initiative (NIfTI) (.nia, .nii, .nii.gz, .hdr, .img, .img.gz)
- Analyze (plain, SPM99, SPM2) (.hdr/.img, .img.gz)
- Digital Imaging and Communications in Medicine (DICOM) (.dcm, .dicom)
- Digital Imaging and Communications in Medicine (DICOM) series (<directory>/)
- Nearly Raw Raster Data (Nrrd) (.nrrd, .nhdr)
- Medical Imaging NetCDF (MINC) (.mnc, .MNC)
- Guys Image Processing Lab (GIPL) (.gipl, .gipl.gz)
Microscopy formats:
- Medical Research Council (MRC) (.mrc, .rec)
- Bio-Rad (.pic, .PIC)
- LSM (Zeiss) microscopy images (.tif, .TIF, .tiff, .TIFF, .lsm, .LSM)
- Stimulate / Signal Data (SDT) (.sdt)
Visualization formats:
- VTK images (.vtk)
Other formats:
- Portable Network Graphics (PNG) (.png, .PNG)
- Joint Photographic Experts Group (JPEG) (.jpg, .JPG, .jpeg, .JPEG)
- Tagged Image File Format (TIFF) (.tif, .TIF, .tiff, .TIFF)
- Windows bitmap (.bmp, .BMP)
- Hierarchical Data Format (HDF5) (.h5 , .hdf5 , .he5)
- MSX-DOS Screen-x (.ge4, .ge5)
MONAI
We’ve arrived at the final one. As they say, “last for best!” The library can be used to implement machine learning algorithms such as image classification and segmentation. MONAI can perform a wide range of operations, from preprocessing data to deploying your final model. I already have some content about it, such as my free 5-hour course on 3D automatic liver segmentation with MONAI.
🆕 NEW
Learn how to effectively manage and process DICOM files in Python with our comprehensive course, designed to equip you with the skills and knowledge you need to succeed.