Introduction
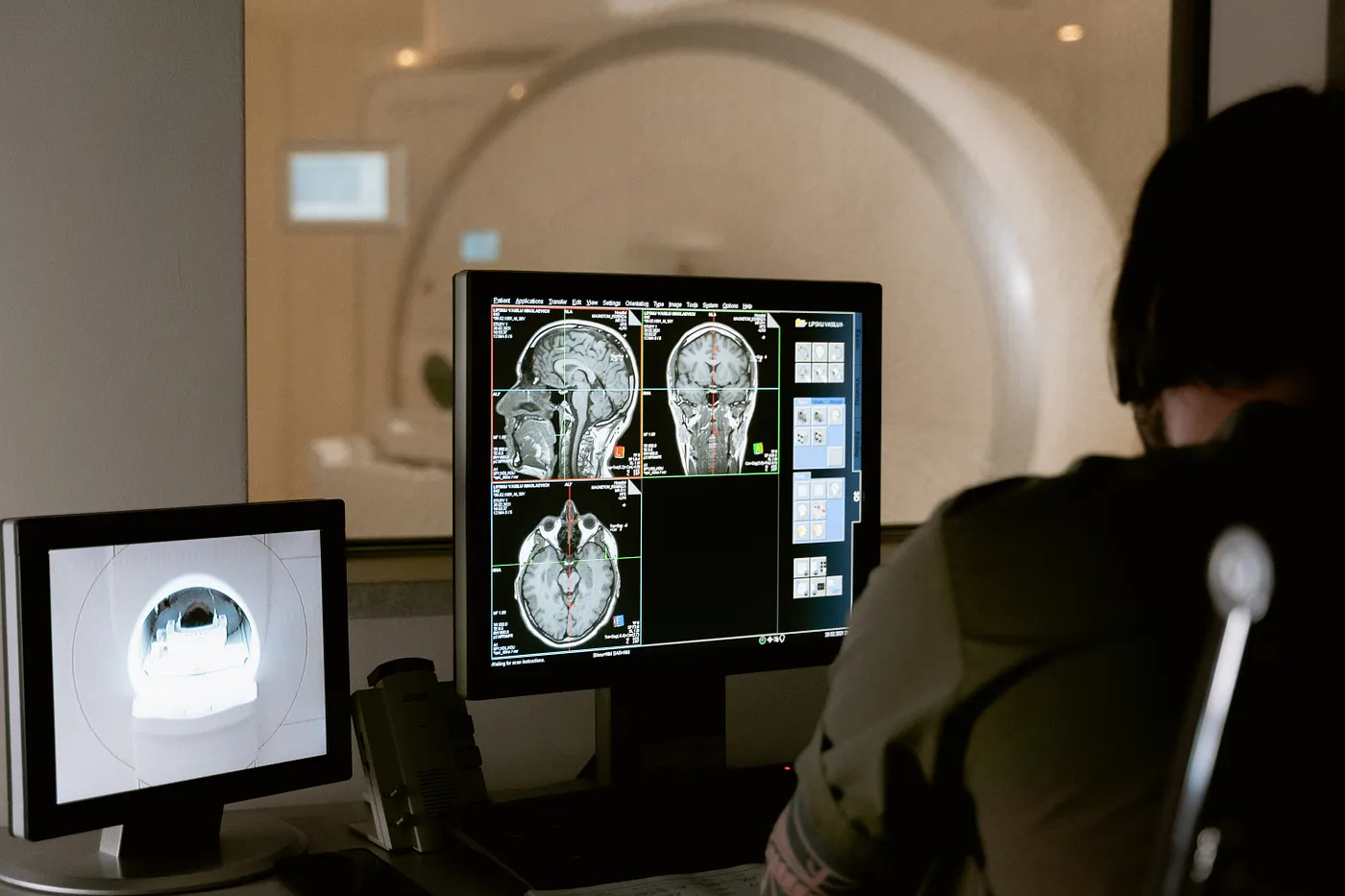
In medical imaging, we use often two types of images, Dicom or Nifti which are the famous ones. But there is a difference between these two types of images or we can say files because they have more information than a normal image. If you want to know these differences then you can see this article.
Now if we want to create a nifti file, means that we will take a set of slices then put them together with the extension “.nii ”.
The process
To do this work you need only two lines of code using python with a library called “dicom2nifti”.
So before starting the code, we need to install the library using the pip installer.
pip install dicom2nifti
After installing the library, we will write the two lines that I told you about, the first one is to import the library and the second one is to use a function that will do all the work.
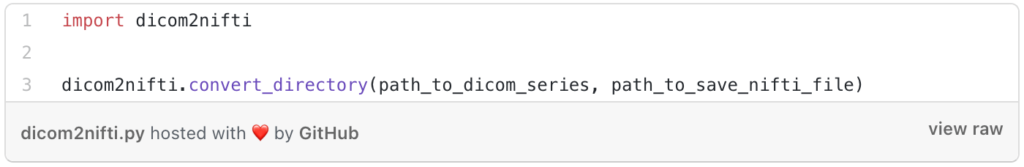
That’s it, by using these two lines you will get your one volume file.
Note: for the path_to_save_nifti_file you should know that you need to include the file name with the extension “.nii ” and if you want a compressed file then you can add “.gz ”.
Convert more than one set of Dicoms
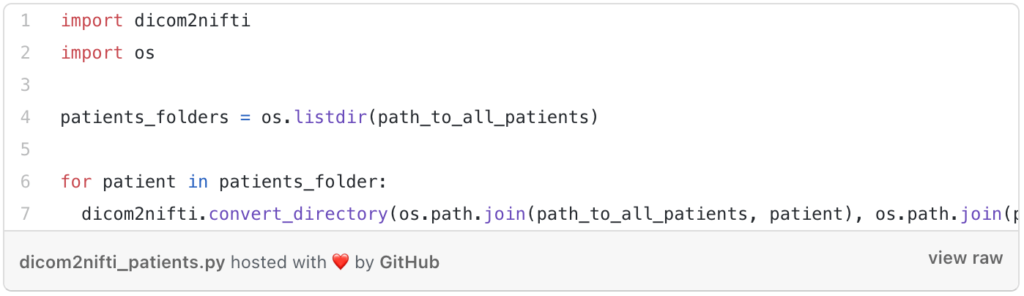
So if you have more than one patient (Dicom series) then you can create a loop that loops in all the patients. It can be something like this:
Explanation
The first step is to list all the patients that you have, then in the loop, we will call the same function “convert_directory” by changing the name of the patient in each step.
Congrats! you will get a nifti file for each patient that you can use to train a deep learning model or something that needs volumetric files.
To visualize the files created, you can use free software like “ITK snap” or “ Slicer 3D” to make sure that you have the right files.