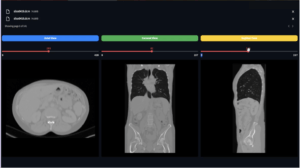
Overview
The DicomMriWindowing module in PYCAD is designed to enhance the contrast and visibility of medical imaging scans, particularly MRI and CBCT scans that exhibit dark contrast. This is achieved through a technique known as windowing, which improves the interpretability of features within the scan by adjusting the image’s intensity scale to focus on specific ranges of interest.
Installation
Ensure that PYCAD is installed and up-to-date. The module requires pydicom and SimpleITK as dependencies, which should be installed in your Python environment.
Usage
Here’s a quick example to get started with DicomMriWindowing:
from pycad.preprocessing import DicomMriWindowing
input_dir = './path/to/input/dicoms'
output_dir = './path/to/save/windowed/dicoms'
windower = DicomMriWindowing(input_dir, output_dir)
windower.window_series()
Class: DicomMriWindowing
Initialization
windower = DicomMriWindowing(input_dir, output_dir)Parameters:
input_dir(str): Path to the directory containing the input DICOM files.output_dir(str): Path where the windowed DICOM files will be saved. The directory is created if it does not exist.
Methods
load_series(dicom_paths)
Loads a series of DICOM images from a specified list of file paths.
Parameters:
dicom_paths(list): A list of file paths to the DICOM images.
Returns:
image_3d(SimpleITK.Image): A 3D SimpleITK image object representing the stacked DICOM series.
apply_windowing(image_3d, coef=4)
Applies intensity windowing to the 3D image to enhance contrast using a mean and standard deviation method.
Parameters:
image_3d(SimpleITK.Image): The 3D image to which windowing will be applied.coef(float, optional): A coefficient determining the spread of the intensity window around the mean intensity. Default is 4.
Returns:
windowed_image(SimpleITK.Image): The windowed image with enhanced contrast.
save_dicom_slice(dcm, slice_arr, index)
Saves a single DICOM slice with the updated pixel data from the windowed image.
Parameters:
dcm(pydicom.Dataset): The original DICOM dataset that corresponds to the slice.slice_arr(numpy.ndarray): The array containing the pixel data for the slice.index(int): The index of the slice within the series.
Returns:
- A boolean indicating whether the save operation was successful.
window_series(coef=4)
Processes an entire series of DICOM images applying windowing and saving the enhanced images.
Parameters:
coef(float, optional): A coefficient for the windowing function. The default is 4.
Theory Behind Windowing
Windowing, also known as level and window, is a technique commonly used in medical imaging to adjust the grayscale values in an image to enhance contrast. By focusing on a specific range of intensity values, structures within the image can be made more or less prominent. This is particularly useful in imaging modalities like MRI or CBCT, where contrast differences are crucial for diagnosis but may be subtle in the raw scan.
Contributing
For any issues, bug reports, or contributions, please open an issue or pull request on the PYCAD GitHub repository.
License
PYCAD is released under the MIT License. For more details, see the LICENSE file.