Keywords: Python, Monai, PyTorch, 3D U-Net, CT scans
Abstract
In this short article, I will discuss the most common issues that you may encounter while using Monai. Because I discovered that sometimes you won’t be able to identify where the errors come from because you won’t be able to find the exact line or function that causes the error in the terminal. As a result, I’ll give you the three errors that took me a long time to figure out how to fix so you don’t waste time like I did.
PS: You will notice that errors are extremely stupid, but you will not realize this until you see the results hahaha.
Errors
- Wrong path to the data
- Wrong keyword in the dictionary
- Multiple classes or pixels values in the segmentation (labels)
Wrong Path to the Data
I know this is a stupid error that you may encounter and that you will say that you can easily find it, but I am afraid to tell you that when you use Monai for the preprocess, it will not tell you that. Especially if you name the folder or the patients incorrectly.
Most of the time, you may encounter an issue with the extension of your data. True, we are using nifti files, so the extension should be ‘.nii,’ but you should be aware that the files may be compressed/zipped, in which case the extension should be ‘.nii.gz,’ and if you forget the ‘.gz,’ the code will not recognize the files that you want to open.
The strange thing here is that it won’t tell you that the directory doesn’t exist or anything, and it won’t even print an error message at the start.
However, when you proceed to the next step, which could be training or even displaying your data, it will inform you that the dataloader is empty.
Note: For this reason, I stated in the previous blog that you should preferably show your patients before the training so that you can correct the paths if you are putting them incorrectly.
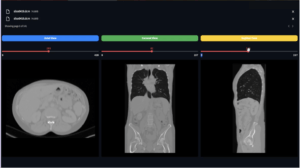
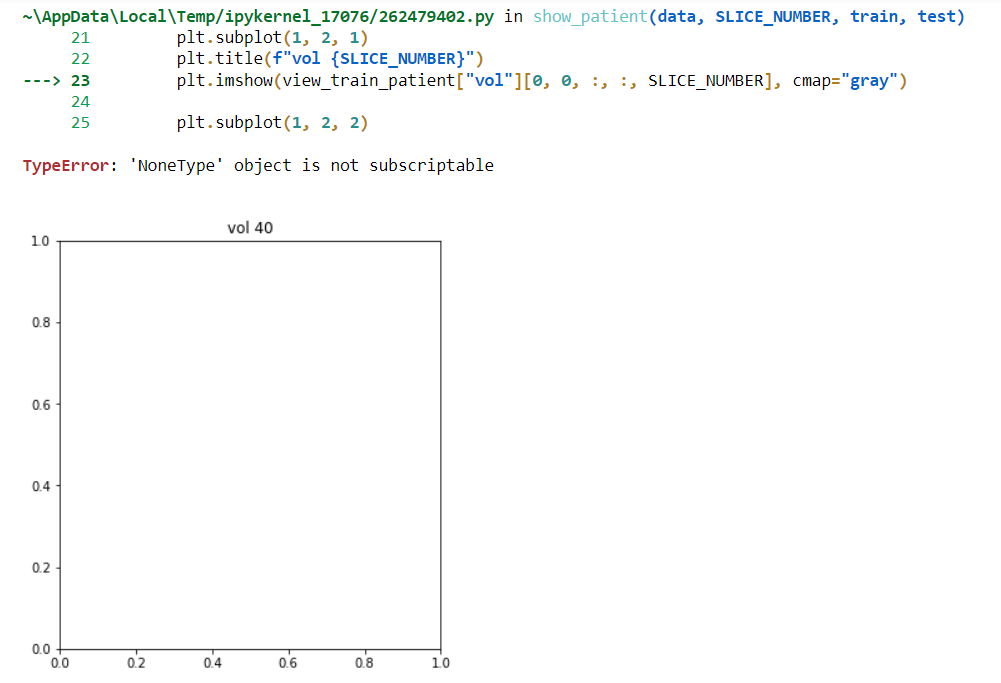
So if you try to show a patient and you get an error like this:
TypeError: 'NoneType' object is not subscriptable
Then you need to verify your paths and files extensions.
Wrong Keyword in the Dictionary
You may misspell one keyword when creating the dictionary to identify the volumes and labels (it happened to me a lot haha). So, of course, it will not work in this case, but fortunately, you will receive an error message indicating that the problem is with the keyword. However, if you make that mistake, you will receive a large text that you will not understand, which may confuse you.
This is how it looks like:
<!-- wp:preformatted -->
<pre class="wp-block-preformatted">=== Transform input info -- Spacingd ===<br><br>=== Transform input info -- Spacingd ===<br><br>=== Transform input info -- Spacingd ===<br><br>=== Transform input info -- Spacingd ===<br><br>=== Transform input info -- Spacingd ===<br>vol statistics:<br>Type: <class 'numpy.ndarray'> float32<br>Shape: (1, 512, 512, 65)<br>Value range: (-1024.0, 1408.0)<br>vol statistics:<br>Type: <class 'numpy.ndarray'> float32<br>Shape: (1, 512, 512, 65)<br>Value range: (-1024.0, 1408.0)<br>vol statistics:<br>Type: <class 'numpy.ndarray'> float32<br>Shape: (1, 512, 512, 65)<br>Value range: (-1024.0, 1408.0)<br>vol statistics:<br>Type: <class 'numpy.ndarray'> float32<br>Shape: (1, 512, 512, 65)<br>Value range: (-1024.0, 1408.0)<br>vol statistics:<br>Type: <class 'numpy.ndarray'> float32<br>Shape: (1, 512, 512, 65)<br>Value range: (-1024.0, 1408.0)<br>seg statistics:<br>Type: <class 'numpy.ndarray'> float32<br>Shape: (1, 512, 512, 65)<br>Value range: (0.0, 1.0)<br>seg statistics:<br>Type: <class 'numpy.ndarray'> float32<br>Shape: (1, 512, 512, 65)<br>Value range: (0.0, 1.0)<br>seg statistics:<br>Type: <class 'numpy.ndarray'> float32<br>Shape: (1, 512, 512, 65)<br>Value range: (0.0, 1.0)<br>seg statistics:<br>Type: <class 'numpy.ndarray'> float32<br>Shape: (1, 512, 512, 65)<br>Value range: (0.0, 1.0)<br>seg statistics:<br>Type: <class 'numpy.ndarray'> float32<br>Shape: (1, 512, 512, 65)<br>Value range: (0.0, 1.0)<br>vol_meta_dict statistics:<br>Type: <class 'dict'> None<br>Value: {'sizeof_hdr': array(348), 'extents': array(0), 'session_error': array(0, dtype=int16), 'dim_info': array(0, dtype=uint8), 'dim': array([ 3, 512, 512, 65, 1, 1, 1, 1], dtype=int16), 'intent_p1': array(0., dtype=float32), 'intent_p2': array(0., dtype=float32), 'intent_p3': array(0., dtype=float32), 'intent_code': array(0, dtype=int16), 'datatype': array(4, dtype=int16), 'bitpix': array(16, dtype=int16), 'slice_start': array(0, dtype=int16), 'pixdim': array([-1. , 0.69921875, 0.69921875, 0.7 , 1. ,<br> 1. , 1. , 1. ], dtype=float32), 'vox_offset': array(0., dtype=float32), 'scl_slope': array(nan, dtype=float32), 'scl_inter': array(nan, dtype=float32), 'slice_end': array(0, dtype=int16), 'slice_code': array(0, dtype=uint8), 'xyzt_units': array(2, dtype=uint8), 'cal_max': array(0., dtype=float32), 'cal_min': array(0., dtype=float32), 'slice_duration': array(0., dtype=float32), 'toffset': array(0., dtype=float32), 'glmax': array(0), 'glmin': array(0), 'qform_code': array(0, dtype=int16), 'sform_code': array(2, dtype=int16), 'quatern_b': array(0., dtype=float32), 'quatern_c': array(1., dtype=float32), 'quatern_d': array(0., dtype=float32), 'qoffset_x': array(169.11578, dtype=float32), 'qoffset_y': array(-320.038, dtype=float32), 'qoffset_z': array(242.8, dtype=float32), 'srow_x': array([ -0.69921875, -0. , 0. , 169.11578 ],<br> dtype=float32), 'srow_y': array([ 0. , 0.69921875, 0. , -320.038 ],<br> dtype=float32), 'srow_z': array([ -0. , 0. , 0.7, 242.8], dtype=float32), 'affine': array([[ -0.69921875, -0. , 0. , 169.11578369],<br> [ 0. , 0.69921875, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'original_affine': array([[ -0.69921875, -0. , 0. , 169.11578369],<br> [ 0. , 0.69921875, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'as_closest_canonical': False, 'spatial_shape': array([512, 512, 65], dtype=int16), 'original_channel_dim': 'no_channel', 'filename_or_obj': 'D:\\Youtube\\Organ and Tumor Segmentation\\datasets\\Task03_Liver\\Data_Train_Test\\TrainVolumes\\p00_5.nii.gz'}<br>vol_meta_dict statistics:<br>Type: <class 'dict'> None<br>Value: {'sizeof_hdr': array(348), 'extents': array(0), 'session_error': array(0, dtype=int16), 'dim_info': array(0, dtype=uint8), 'dim': array([ 3, 512, 512, 65, 1, 1, 1, 1], dtype=int16), 'intent_p1': array(0., dtype=float32), 'intent_p2': array(0., dtype=float32), 'intent_p3': array(0., dtype=float32), 'intent_code': array(0, dtype=int16), 'datatype': array(4, dtype=int16), 'bitpix': array(16, dtype=int16), 'slice_start': array(0, dtype=int16), 'pixdim': array([-1. , 0.69921875, 0.69921875, 0.7 , 1. ,<br> 1. , 1. , 1. ], dtype=float32), 'vox_offset': array(0., dtype=float32), 'scl_slope': array(nan, dtype=float32), 'scl_inter': array(nan, dtype=float32), 'slice_end': array(0, dtype=int16), 'slice_code': array(0, dtype=uint8), 'xyzt_units': array(2, dtype=uint8), 'cal_max': array(0., dtype=float32), 'cal_min': array(0., dtype=float32), 'slice_duration': array(0., dtype=float32), 'toffset': array(0., dtype=float32), 'glmax': array(0), 'glmin': array(0), 'qform_code': array(0, dtype=int16), 'sform_code': array(2, dtype=int16), 'quatern_b': array(0., dtype=float32), 'quatern_c': array(1., dtype=float32), 'quatern_d': array(0., dtype=float32), 'qoffset_x': array(169.11578, dtype=float32), 'qoffset_y': array(-320.038, dtype=float32), 'qoffset_z': array(242.8, dtype=float32), 'srow_x': array([ -0.69921875, -0. , 0. , 169.11578 ],<br> dtype=float32), 'srow_y': array([ 0. , 0.69921875, 0. , -320.038 ],<br> dtype=float32), 'srow_z': array([ -0. , 0. , 0.7, 242.8], dtype=float32), 'affine': array([[ -0.69921875, -0. , 0. , 169.11578369],<br> [ 0. , 0.69921875, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'original_affine': array([[ -0.69921875, -0. , 0. , 169.11578369],<br> [ 0. , 0.69921875, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'as_closest_canonical': False, 'spatial_shape': array([512, 512, 65], dtype=int16), 'original_channel_dim': 'no_channel', 'filename_or_obj': 'D:\\Youtube\\Organ and Tumor Segmentation\\datasets\\Task03_Liver\\Data_Train_Test\\TrainVolumes\\p00_5.nii.gz'}<br>vol_meta_dict statistics:<br>Type: <class 'dict'> None<br>Value: {'sizeof_hdr': array(348), 'extents': array(0), 'session_error': array(0, dtype=int16), 'dim_info': array(0, dtype=uint8), 'dim': array([ 3, 512, 512, 65, 1, 1, 1, 1], dtype=int16), 'intent_p1': array(0., dtype=float32), 'intent_p2': array(0., dtype=float32), 'intent_p3': array(0., dtype=float32), 'intent_code': array(0, dtype=int16), 'datatype': array(4, dtype=int16), 'bitpix': array(16, dtype=int16), 'slice_start': array(0, dtype=int16), 'pixdim': array([-1. , 0.69921875, 0.69921875, 0.7 , 1. ,<br> 1. , 1. , 1. ], dtype=float32), 'vox_offset': array(0., dtype=float32), 'scl_slope': array(nan, dtype=float32), 'scl_inter': array(nan, dtype=float32), 'slice_end': array(0, dtype=int16), 'slice_code': array(0, dtype=uint8), 'xyzt_units': array(2, dtype=uint8), 'cal_max': array(0., dtype=float32), 'cal_min': array(0., dtype=float32), 'slice_duration': array(0., dtype=float32), 'toffset': array(0., dtype=float32), 'glmax': array(0), 'glmin': array(0), 'qform_code': array(0, dtype=int16), 'sform_code': array(2, dtype=int16), 'quatern_b': array(0., dtype=float32), 'quatern_c': array(1., dtype=float32), 'quatern_d': array(0., dtype=float32), 'qoffset_x': array(169.11578, dtype=float32), 'qoffset_y': array(-320.038, dtype=float32), 'qoffset_z': array(242.8, dtype=float32), 'srow_x': array([ -0.69921875, -0. , 0. , 169.11578 ],<br> dtype=float32), 'srow_y': array([ 0. , 0.69921875, 0. , -320.038 ],<br> dtype=float32), 'srow_z': array([ -0. , 0. , 0.7, 242.8], dtype=float32), 'affine': array([[ -0.69921875, -0. , 0. , 169.11578369],<br> [ 0. , 0.69921875, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'original_affine': array([[ -0.69921875, -0. , 0. , 169.11578369],<br> [ 0. , 0.69921875, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'as_closest_canonical': False, 'spatial_shape': array([512, 512, 65], dtype=int16), 'original_channel_dim': 'no_channel', 'filename_or_obj': 'D:\\Youtube\\Organ and Tumor Segmentation\\datasets\\Task03_Liver\\Data_Train_Test\\TrainVolumes\\p00_5.nii.gz'}</pre>
<!-- /wp:preformatted -->
<!-- wp:preformatted -->
<pre class="wp-block-preformatted">vol_meta_dict statistics:<br>Type: <class 'dict'> None<br>Value: {'sizeof_hdr': array(348), 'extents': array(0), 'session_error': array(0, dtype=int16), 'dim_info': array(0, dtype=uint8), 'dim': array([ 3, 512, 512, 65, 1, 1, 1, 1], dtype=int16), 'intent_p1': array(0., dtype=float32), 'intent_p2': array(0., dtype=float32), 'intent_p3': array(0., dtype=float32), 'intent_code': array(0, dtype=int16), 'datatype': array(4, dtype=int16), 'bitpix': array(16, dtype=int16), 'slice_start': array(0, dtype=int16), 'pixdim': array([-1. , 0.69921875, 0.69921875, 0.7 , 1. ,<br> 1. , 1. , 1. ], dtype=float32), 'vox_offset': array(0., dtype=float32), 'scl_slope': array(nan, dtype=float32), 'scl_inter': array(nan, dtype=float32), 'slice_end': array(0, dtype=int16), 'slice_code': array(0, dtype=uint8), 'xyzt_units': array(2, dtype=uint8), 'cal_max': array(0., dtype=float32), 'cal_min': array(0., dtype=float32), 'slice_duration': array(0., dtype=float32), 'toffset': array(0., dtype=float32), 'glmax': array(0), 'glmin': array(0), 'qform_code': array(0, dtype=int16), 'sform_code': array(2, dtype=int16), 'quatern_b': array(0., dtype=float32), 'quatern_c': array(1., dtype=float32), 'quatern_d': array(0., dtype=float32), 'qoffset_x': array(169.11578, dtype=float32), 'qoffset_y': array(-320.038, dtype=float32), 'qoffset_z': array(242.8, dtype=float32), 'srow_x': array([ -0.69921875, -0. , 0. , 169.11578 ],<br> dtype=float32), 'srow_y': array([ 0. , 0.69921875, 0. , -320.038 ],<br> dtype=float32), 'srow_z': array([ -0. , 0. , 0.7, 242.8], dtype=float32), 'affine': array([[ -0.69921875, -0. , 0. , 169.11578369],<br> [ 0. , 0.69921875, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'original_affine': array([[ -0.69921875, -0. , 0. , 169.11578369],<br> [ 0. , 0.69921875, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'as_closest_canonical': False, 'spatial_shape': array([512, 512, 65], dtype=int16), 'original_channel_dim': 'no_channel', 'filename_or_obj': 'D:\\Youtube\\Organ and Tumor Segmentation\\datasets\\Task03_Liver\\Data_Train_Test\\TrainVolumes\\p00_5.nii.gz'}<br>vol_meta_dict statistics:<br>Type: <class 'dict'> None<br>Value: {'sizeof_hdr': array(348), 'extents': array(0), 'session_error': array(0, dtype=int16), 'dim_info': array(0, dtype=uint8), 'dim': array([ 3, 512, 512, 65, 1, 1, 1, 1], dtype=int16), 'intent_p1': array(0., dtype=float32), 'intent_p2': array(0., dtype=float32), 'intent_p3': array(0., dtype=float32), 'intent_code': array(0, dtype=int16), 'datatype': array(4, dtype=int16), 'bitpix': array(16, dtype=int16), 'slice_start': array(0, dtype=int16), 'pixdim': array([-1. , 0.69921875, 0.69921875, 0.7 , 1. ,<br> 1. , 1. , 1. ], dtype=float32), 'vox_offset': array(0., dtype=float32), 'scl_slope': array(nan, dtype=float32), 'scl_inter': array(nan, dtype=float32), 'slice_end': array(0, dtype=int16), 'slice_code': array(0, dtype=uint8), 'xyzt_units': array(2, dtype=uint8), 'cal_max': array(0., dtype=float32), 'cal_min': array(0., dtype=float32), 'slice_duration': array(0., dtype=float32), 'toffset': array(0., dtype=float32), 'glmax': array(0), 'glmin': array(0), 'qform_code': array(0, dtype=int16), 'sform_code': array(2, dtype=int16), 'quatern_b': array(0., dtype=float32), 'quatern_c': array(1., dtype=float32), 'quatern_d': array(0., dtype=float32), 'qoffset_x': array(169.11578, dtype=float32), 'qoffset_y': array(-320.038, dtype=float32), 'qoffset_z': array(242.8, dtype=float32), 'srow_x': array([ -0.69921875, -0. , 0. , 169.11578 ],<br> dtype=float32), 'srow_y': array([ 0. , 0.69921875, 0. , -320.038 ],<br> dtype=float32), 'srow_z': array([ -0. , 0. , 0.7, 242.8], dtype=float32), 'affine': array([[ -0.69921875, -0. , 0. , 169.11578369],<br> [ 0. , 0.69921875, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'original_affine': array([[ -0.69921875, -0. , 0. , 169.11578369],<br> [ 0. , 0.69921875, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'as_closest_canonical': False, 'spatial_shape': array([512, 512, 65], dtype=int16), 'original_channel_dim': 'no_channel', 'filename_or_obj': 'D:\\Youtube\\Organ and Tumor Segmentation\\datasets\\Task03_Liver\\Data_Train_Test\\TrainVolumes\\p00_5.nii.gz'}<br>seg_meta_dict statistics:<br>Type: <class 'dict'> None<br>Value: {'sizeof_hdr': array(348), 'extents': array(0), 'session_error': array(0, dtype=int16), 'dim_info': array(0, dtype=uint8), 'dim': array([ 3, 512, 512, 65, 1, 1, 1, 1], dtype=int16), 'intent_p1': array(0., dtype=float32), 'intent_p2': array(0., dtype=float32), 'intent_p3': array(0., dtype=float32), 'intent_code': array(0, dtype=int16), 'datatype': array(2, dtype=int16), 'bitpix': array(8, dtype=int16), 'slice_start': array(0, dtype=int16), 'pixdim': array([-1. , 0.6992188, 0.6992188, 0.7 , 1. ,<br> 1. , 1. , 1. ], dtype=float32), 'vox_offset': array(0., dtype=float32), 'scl_slope': array(nan, dtype=float32), 'scl_inter': array(nan, dtype=float32), 'slice_end': array(0, dtype=int16), 'slice_code': array(0, dtype=uint8), 'xyzt_units': array(2, dtype=uint8), 'cal_max': array(0., dtype=float32), 'cal_min': array(0., dtype=float32), 'slice_duration': array(0., dtype=float32), 'toffset': array(0., dtype=float32), 'glmax': array(0), 'glmin': array(0), 'qform_code': array(0, dtype=int16), 'sform_code': array(2, dtype=int16), 'quatern_b': array(0., dtype=float32), 'quatern_c': array(1., dtype=float32), 'quatern_d': array(0., dtype=float32), 'qoffset_x': array(169.11581, dtype=float32), 'qoffset_y': array(-320.038, dtype=float32), 'qoffset_z': array(242.8, dtype=float32), 'srow_x': array([ -0.6992188, -0. , 0. , 169.11581 ], dtype=float32), 'srow_y': array([ 0. , 0.6992188, 0. , -320.038 ],<br> dtype=float32), 'srow_z': array([ -0. , 0. , 0.7, 242.8], dtype=float32), 'affine': array([[ -0.69921881, -0. , 0. , 169.11581421],<br> [ 0. , 0.69921881, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'original_affine': array([[ -0.69921881, -0. , 0. , 169.11581421],<br> [ 0. , 0.69921881, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'as_closest_canonical': False, 'spatial_shape': array([512, 512, 65], dtype=int16), 'original_channel_dim': 'no_channel', 'filename_or_obj': 'D:\\Youtube\\Organ and Tumor Segmentation\\datasets\\Task03_Liver\\Data_Train_Test\\TrainSegmentation\\p00_5.nii.gz'}<br>seg_meta_dict statistics:<br>Type: <class 'dict'> None<br>Value: {'sizeof_hdr': array(348), 'extents': array(0), 'session_error': array(0, dtype=int16), 'dim_info': array(0, dtype=uint8), 'dim': array([ 3, 512, 512, 65, 1, 1, 1, 1], dtype=int16), 'intent_p1': array(0., dtype=float32), 'intent_p2': array(0., dtype=float32), 'intent_p3': array(0., dtype=float32), 'intent_code': array(0, dtype=int16), 'datatype': array(2, dtype=int16), 'bitpix': array(8, dtype=int16), 'slice_start': array(0, dtype=int16), 'pixdim': array([-1. , 0.6992188, 0.6992188, 0.7 , 1. ,<br> 1. , 1. , 1. ], dtype=float32), 'vox_offset': array(0., dtype=float32), 'scl_slope': array(nan, dtype=float32), 'scl_inter': array(nan, dtype=float32), 'slice_end': array(0, dtype=int16), 'slice_code': array(0, dtype=uint8), 'xyzt_units': array(2, dtype=uint8), 'cal_max': array(0., dtype=float32), 'cal_min': array(0., dtype=float32), 'slice_duration': array(0., dtype=float32), 'toffset': array(0., dtype=float32), 'glmax': array(0), 'glmin': array(0), 'qform_code': array(0, dtype=int16), 'sform_code': array(2, dtype=int16), 'quatern_b': array(0., dtype=float32), 'quatern_c': array(1., dtype=float32), 'quatern_d': array(0., dtype=float32), 'qoffset_x': array(169.11581, dtype=float32), 'qoffset_y': array(-320.038, dtype=float32), 'qoffset_z': array(242.8, dtype=float32), 'srow_x': array([ -0.6992188, -0. , 0. , 169.11581 ], dtype=float32), 'srow_y': array([ 0. , 0.6992188, 0. , -320.038 ],<br> dtype=float32), 'srow_z': array([ -0. , 0. , 0.7, 242.8], dtype=float32), 'affine': array([[ -0.69921881, -0. , 0. , 169.11581421],<br> [ 0. , 0.69921881, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'original_affine': array([[ -0.69921881, -0. , 0. , 169.11581421],<br> [ 0. , 0.69921881, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'as_closest_canonical': False, 'spatial_shape': array([512, 512, 65], dtype=int16), 'original_channel_dim': 'no_channel', 'filename_or_obj': 'D:\\Youtube\\Organ and Tumor Segmentation\\datasets\\Task03_Liver\\Data_Train_Test\\TrainSegmentation\\p00_5.nii.gz'}</pre>
<!-- /wp:preformatted -->
<!-- wp:preformatted -->
<pre class="wp-block-preformatted">seg_meta_dict statistics:<br>Type: <class 'dict'> None<br>Value: {'sizeof_hdr': array(348), 'extents': array(0), 'session_error': array(0, dtype=int16), 'dim_info': array(0, dtype=uint8), 'dim': array([ 3, 512, 512, 65, 1, 1, 1, 1], dtype=int16), 'intent_p1': array(0., dtype=float32), 'intent_p2': array(0., dtype=float32), 'intent_p3': array(0., dtype=float32), 'intent_code': array(0, dtype=int16), 'datatype': array(2, dtype=int16), 'bitpix': array(8, dtype=int16), 'slice_start': array(0, dtype=int16), 'pixdim': array([-1. , 0.6992188, 0.6992188, 0.7 , 1. ,<br> 1. , 1. , 1. ], dtype=float32), 'vox_offset': array(0., dtype=float32), 'scl_slope': array(nan, dtype=float32), 'scl_inter': array(nan, dtype=float32), 'slice_end': array(0, dtype=int16), 'slice_code': array(0, dtype=uint8), 'xyzt_units': array(2, dtype=uint8), 'cal_max': array(0., dtype=float32), 'cal_min': array(0., dtype=float32), 'slice_duration': array(0., dtype=float32), 'toffset': array(0., dtype=float32), 'glmax': array(0), 'glmin': array(0), 'qform_code': array(0, dtype=int16), 'sform_code': array(2, dtype=int16), 'quatern_b': array(0., dtype=float32), 'quatern_c': array(1., dtype=float32), 'quatern_d': array(0., dtype=float32), 'qoffset_x': array(169.11581, dtype=float32), 'qoffset_y': array(-320.038, dtype=float32), 'qoffset_z': array(242.8, dtype=float32), 'srow_x': array([ -0.6992188, -0. , 0. , 169.11581 ], dtype=float32), 'srow_y': array([ 0. , 0.6992188, 0. , -320.038 ],<br> dtype=float32), 'srow_z': array([ -0. , 0. , 0.7, 242.8], dtype=float32), 'affine': array([[ -0.69921881, -0. , 0. , 169.11581421],<br> [ 0. , 0.69921881, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'original_affine': array([[ -0.69921881, -0. , 0. , 169.11581421],<br> [ 0. , 0.69921881, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'as_closest_canonical': False, 'spatial_shape': array([512, 512, 65], dtype=int16), 'original_channel_dim': 'no_channel', 'filename_or_obj': 'D:\\Youtube\\Organ and Tumor Segmentation\\datasets\\Task03_Liver\\Data_Train_Test\\TrainSegmentation\\p00_5.nii.gz'}<br>seg_meta_dict statistics:<br>Type: <class 'dict'> None<br>Value: {'sizeof_hdr': array(348), 'extents': array(0), 'session_error': array(0, dtype=int16), 'dim_info': array(0, dtype=uint8), 'dim': array([ 3, 512, 512, 65, 1, 1, 1, 1], dtype=int16), 'intent_p1': array(0., dtype=float32), 'intent_p2': array(0., dtype=float32), 'intent_p3': array(0., dtype=float32), 'intent_code': array(0, dtype=int16), 'datatype': array(2, dtype=int16), 'bitpix': array(8, dtype=int16), 'slice_start': array(0, dtype=int16), 'pixdim': array([-1. , 0.6992188, 0.6992188, 0.7 , 1. ,<br> 1. , 1. , 1. ], dtype=float32), 'vox_offset': array(0., dtype=float32), 'scl_slope': array(nan, dtype=float32), 'scl_inter': array(nan, dtype=float32), 'slice_end': array(0, dtype=int16), 'slice_code': array(0, dtype=uint8), 'xyzt_units': array(2, dtype=uint8), 'cal_max': array(0., dtype=float32), 'cal_min': array(0., dtype=float32), 'slice_duration': array(0., dtype=float32), 'toffset': array(0., dtype=float32), 'glmax': array(0), 'glmin': array(0), 'qform_code': array(0, dtype=int16), 'sform_code': array(2, dtype=int16), 'quatern_b': array(0., dtype=float32), 'quatern_c': array(1., dtype=float32), 'quatern_d': array(0., dtype=float32), 'qoffset_x': array(169.11581, dtype=float32), 'qoffset_y': array(-320.038, dtype=float32), 'qoffset_z': array(242.8, dtype=float32), 'srow_x': array([ -0.6992188, -0. , 0. , 169.11581 ], dtype=float32), 'srow_y': array([ 0. , 0.6992188, 0. , -320.038 ],<br> dtype=float32), 'srow_z': array([ -0. , 0. , 0.7, 242.8], dtype=float32), 'affine': array([[ -0.69921881, -0. , 0. , 169.11581421],<br> [ 0. , 0.69921881, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'original_affine': array([[ -0.69921881, -0. , 0. , 169.11581421],<br> [ 0. , 0.69921881, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'as_closest_canonical': False, 'spatial_shape': array([512, 512, 65], dtype=int16), 'original_channel_dim': 'no_channel', 'filename_or_obj': 'D:\\Youtube\\Organ and Tumor Segmentation\\datasets\\Task03_Liver\\Data_Train_Test\\TrainSegmentation\\p00_5.nii.gz'}<br>seg_meta_dict statistics:<br>Type: <class 'dict'> None<br>Value: {'sizeof_hdr': array(348), 'extents': array(0), 'session_error': array(0, dtype=int16), 'dim_info': array(0, dtype=uint8), 'dim': array([ 3, 512, 512, 65, 1, 1, 1, 1], dtype=int16), 'intent_p1': array(0., dtype=float32), 'intent_p2': array(0., dtype=float32), 'intent_p3': array(0., dtype=float32), 'intent_code': array(0, dtype=int16), 'datatype': array(2, dtype=int16), 'bitpix': array(8, dtype=int16), 'slice_start': array(0, dtype=int16), 'pixdim': array([-1. , 0.6992188, 0.6992188, 0.7 , 1. ,<br> 1. , 1. , 1. ], dtype=float32), 'vox_offset': array(0., dtype=float32), 'scl_slope': array(nan, dtype=float32), 'scl_inter': array(nan, dtype=float32), 'slice_end': array(0, dtype=int16), 'slice_code': array(0, dtype=uint8), 'xyzt_units': array(2, dtype=uint8), 'cal_max': array(0., dtype=float32), 'cal_min': array(0., dtype=float32), 'slice_duration': array(0., dtype=float32), 'toffset': array(0., dtype=float32), 'glmax': array(0), 'glmin': array(0), 'qform_code': array(0, dtype=int16), 'sform_code': array(2, dtype=int16), 'quatern_b': array(0., dtype=float32), 'quatern_c': array(1., dtype=float32), 'quatern_d': array(0., dtype=float32), 'qoffset_x': array(169.11581, dtype=float32), 'qoffset_y': array(-320.038, dtype=float32), 'qoffset_z': array(242.8, dtype=float32), 'srow_x': array([ -0.6992188, -0. , 0. , 169.11581 ], dtype=float32), 'srow_y': array([ 0. , 0.6992188, 0. , -320.038 ],<br> dtype=float32), 'srow_z': array([ -0. , 0. , 0.7, 242.8], dtype=float32), 'affine': array([[ -0.69921881, -0. , 0. , 169.11581421],<br> [ 0. , 0.69921881, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'original_affine': array([[ -0.69921881, -0. , 0. , 169.11581421],<br> [ 0. , 0.69921881, 0. , -320.03799438],<br> [ -0. , 0. , 0.69999999, 242.80000305],<br> [ 0. , 0. , 0. , 1. ]]), 'as_closest_canonical': False, 'spatial_shape': array([512, 512, 65], dtype=int16), 'original_channel_dim': 'no_channel', 'filename_or_obj': 'D:\\Youtube\\Organ and Tumor Segmentation\\datasets\\Task03_Liver\\Data_Train_Test\\TrainSegmentation\\p00_5.nii.gz'}</pre>
<!-- /wp:preformatted -->But you don’t need to know what they are putting here, because nothing is important.
When you encounter this type of error, simply go to the bottom of the error and you will see something like this:
RuntimeError: applying transform <monai.transforms.compose.Compose object at 0x000001A4A682AC10>
Or
KeyError: 'Key was missing (voll) and allow_missing_keys==False'
Or
RuntimeError: applying transform <monai.transforms.spatial.dictionary.Spacingd object at 0x000001A4A682A250>
So, if you get these kinds of errors, you should double-check the keywords you’re using in the dictionary :).
Multiple Classes or Pixels Values in the Segmentation (labels)
And this is the most common error that leaves people stranded for hours, if not days. Because you will never understand the type of errors unless you have already encountered them or someone assists you in learning about them.
This may be a problem if you download a public dataset or if you have a labeled dataset but don’t know who labeled it. When people segment, they may segment for a different task than what you’re trying to do, or they may make mistakes that they don’t notice at first.
As an example, suppose you’re attempting to do liver segmentation and you download a dataset that has previously been used for liver and liver tumor segmentation. The labels in this case will have three different values (classes): background, liver, and tumor. As a result, if you use the same dataset without making any changes, it will not work. Because you will specify in your code that you only have two classes (background and liver), and when the code runs the data, it will find three different values that will automatically place them in three classes. So you’ll get a Cuda error, and you’ll think the problem is with your GPU or Cuda installation, but it’s actually with the data you’re using.
RuntimeError: 'CUDA error: device-side assert triggered'
So, if you get this type of error, you should double-check your data. If you want to learn more about this error, check out this blog post.
What next!
In the following blog post, I’ll show you how to run the training on your machine.
Please keep in mind that I will be using the same code as Monai, with a few minor changes.