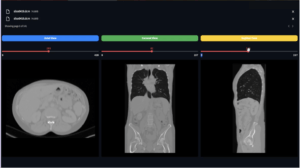
Overview
The NiftiCTWindowing module is an integral part of the PYCAD library designed to enhance the visual quality of CT (Computed Tomography) images using the technique of windowing. This documentation explains the theoretical background, usage, and functions provided by the NiftiCTWindowing class within the module.
What is Windowing?
Windowing, also known as grey-level mapping, contrast stretching, or histogram modification, is a technique used in medical imaging to improve the visibility of certain structures in CT scans. In CT imaging, windowing is essential because the range of intensities (or Hounsfield units) captured by a scanner is much broader than what can be displayed on a standard monitor or interpreted by the human eye.
A windowing operation involves selecting a range of intensities (the window width) around a central intensity value (the window level or center) and mapping this range to the displayable range of intensities (usually 0-255 for an 8-bit display). Intensities outside the selected range are either set to the minimum or maximum displayable value. This enhances the contrast within the specified window and allows radiologists to focus on particular tissues such as soft tissue, lung tissue, or bone.
Class NiftiCTWindowing
The NiftiCTWindowing class provides an interface to apply windowing operations on NIfTI-format MRI images. It is specifically designed to handle CT images, which typically require careful adjustment of window levels to accurately visualize different tissue densities.
Constructor Parameters
window_center: The center of the windowing operation (default 40), representing the midpoint of the range of intensities of interest.window_width: The width of the windowing operation (default 400), defining the size of the range around the window center to be mapped to the displayable intensity range.visualize: A boolean flag indicating whether to visualize an example slice before and after windowing (default False).
Methods:
apply_windowing(image)
Applies the windowing operation to an input image
- Parameters: A single NIfTI image array.
- Returns: A windowed image array, with enhanced contrast based on the specified window center and width.
process_file(nifti_path, output_path)
Processes a single NIfTI file applying windowing and saving the result.
- Parameters:
nifti_path: The file path of the input NIfTI image.output_path: The file path where the windowed NIfTI image should be saved.
- Returns: A tuple containing the original and the windowed image arrays.
convert(nifti_path, output_path)
Converts an input NIfTI file using the windowing parameters provided upon instantiation and saves the windowed image to the specified output path. Optionally, it can display an example slice before and after windowing if the visualize parameter is set to True.
- Parameters:
nifti_path: The file path of the input NIfTI image.output_path: The file path where the windowed NIfTI image should be saved.
display_example_slice(original_image, windowed_image)
Displays a side-by-side comparison of the original and windowed images for a selected slice, typically the middle slice in the 3D volume. This method is invoked when the visualize the parameter is True during the convert operation.
Usage Example:
from pycad.preprocessing import NiftiCTWindowing
# Paths for the input and output files
nifti_path = 'path/to/your/input/nifti.nii.gz'
output_path = 'path/to/save/windowed_nifti.nii.gz'
# Instantiate the windowing class with custom parameters
windower = NiftiCTWindowing(window_center=40, window_width=400, visualize=True)
# Apply windowing and visualize the result
windower.convert(nifti_path, output_path)
Notes:
- Ensure the input NIfTI images are correctly scaled in Hounsfield units for accurate windowing.
- The default parameters (window_center=40, window_width=400) are typically suitable for visualizing soft tissue. Adjust them based on the specific tissue or structure of interest.
- The visualization feature is helpful for quality assurance and to confirm that the windowing parameters are correctly set for the intended purpose.
Conclusion
The NiftiCTWindowing module in PYCAD is a powerful tool for medical imaging professionals and researchers working with CT data. It streamlines the process of enhancing the visual quality of CT images, allowing for more accurate and efficient analysis.